Abstract
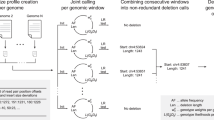
Recent work has shown that copy number polymorphism is an important class of genetic variation in human genomes1,2,3,4. Here we report a new method that uses SNP genotype data from parent-offspring trios to identify polymorphic deletions. We applied this method to data from the International HapMap Project5 to produce the first high-resolution population surveys of deletion polymorphism. Approximately 100 of these deletions have been experimentally validated using comparative genome hybridization on tiling-resolution oligonucleotide microarrays. Our analysis identifies a total of 586 distinct regions that harbor deletion polymorphisms in one or more of the families. Notably, we estimate that typical individuals are hemizygous for roughly 30–50 deletions larger than 5 kb, totaling around 550–750 kb of euchromatic sequence across their genomes. The detected deletions span a total of 267 known and predicted genes. Overall, however, the deleted regions are relatively gene-poor, consistent with the action of purifying selection against deletions. Deletion polymorphisms may well have an important role in the genetics of complex traits; however, they are not directly observed in most current gene mapping studies. Our new method will permit the identification of deletion polymorphisms in high-density SNP surveys of trio or other family data.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Sebat, J. et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004).
Iafrate, A.J. et al. Detection of large-scale variation in the human genome. Nat. Genet. 36, 949–951 (2004).
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nat. Genet. 37, 727–732 (2005).
Sharp, A.J. et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77, 78–88 (2005).
The International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
Schmickel, R.D. Contiguous gene syndromes: a component of recognizable syndromes. J. Pediatr. 109, 231–241 (1986).
Chen, K.S. et al. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat. Genet. 17, 154–163 (1997).
Flint, J. et al. The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat. Genet. 9, 132–140 (1995).
Gardner, R.J. & Sutherland, G.R. Chromosomes Abnormalities and Genetic Counseling (Oxford Univ. Press, Oxford, 2004).
Yu, C.E. et al. Presence of large deletions in kindreds with autism. Am. J. Hum. Genet. 71, 100–115 (2002).
Petrov, D.A. Mutational equilibrium model of genome size evolution. Theor. Popul. Biol. 61, 531–544 (2002).
Olson, M.V. When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64, 18–23 (1999).
Amos, C.I., Shete, S., Chen, J. & Yu, R.K. Positional identification of microdeletions with genetic markers. Hum. Hered. 56, 107–118 (2003).
Giglio, S. et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 68, 874–883 (2001).
Weber, J.L. et al. Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet. 71, 854–862 (2002).
Bhangale, T.R., Rieder, M.J., Livingston, R.J. & Nickerson, D.A. Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum. Mol. Genet. 14, 59–69 (2005).
Carter, N.P. As normal as normal can be? Nat. Genet. 36, 931–932 (2004).
Bailey, J.A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002).
Clark, A.G. et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 302, 1960–1963 (2003).
Emes, R.D., Goodstadt, L., Winter, E.E. & Ponting, C.P. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum. Mol. Genet. 12, 701–709 (2003).
Hinds, D.A., Kloek, A.P. & Frazer, K.A. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. Advance online publication, 4 December 2005 (10.1038/ng1695).
McCarroll, S.A. et al. Common deletion polymorphisms in the human genome. Nat. Genet. Advance online publication, 4 December 2005 (10.1038/ng1696).
Fredman, D. et al. Complex SNP-related sequence variation in segmental genome duplications. Nat. Genet. 36, 861–866 (2004).
Selzer, R.R. et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosom. Cancer 44, 305–319 (2005).
Acknowledgements
We thank G. Coop, D. Cutler, A. DiRienzo, H. Fiegler, M. Przeworski, G. Raca, C. Tyler-Smith and D. Vetrie for comments and discussions; R. Redon for managing the genomic DNA collection; B. Voight for extracting the SNP classifications; S. Das, A. DiRienzo and C. Ober for lab space and equipment and the members of the International HapMap Consortium for their work in creating this dataset. This work was supported by a grant to J.K.P. from the Packard Foundation. D.F.C. was supported in part by US National Institutes of Health/National Institute of General Medical Sciences Genetics and Regulation Training Grant GM07197. Additional funding was provided by the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
List of deletions. (XLS 48 kb)
Supplementary Table 2
List of deletions. (XLS 72 kb)
Supplementary Table 3
List of CGH validation results. (XLS 48 kb)
Supplementary Table 4
List of genes involved. (XLS 90 kb)
Rights and permissions
About this article
Cite this article
Conrad, D., Andrews, T., Carter, N. et al. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 38, 75–81 (2006). https://doi.org/10.1038/ng1697
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1697
This article is cited by
-
Assessment of linkage disequilibrium patterns between structural variants and single nucleotide polymorphisms in three commercial chicken populations
BMC Genomics (2022)
-
Genome-wide detection of CNVs and their association with performance traits in broilers
BMC Genomics (2021)
-
Benchmarking germline CNV calling tools from exome sequencing data
Scientific Reports (2021)
-
Identification and population genetic analyses of copy number variations in six domestic goat breeds and Bezoar ibexes using next-generation sequencing
BMC Genomics (2020)
-
Copy number variation is highly correlated with differential gene expression: a pan-cancer study
BMC Medical Genetics (2019)