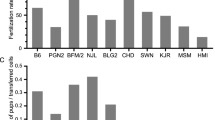
Abstract
The mouse is the foremost vertebrate experimental model because its genome can be precisely and variously engineered. Now that the mouse genome has been sequenced and annotated, the task of mutating each gene is feasible, and an international cooperation is providing mutated embryonic stem cells and mice as readily available resources. Because these resources will change biomedical research, decisions about their nature will have far-reaching effects. It is therefore timely to consider topical issues for mouse genome engineering, such as the background genotype; homologous, site-specific and transpositional recombination; conditional mutagenesis; RNA-mediated interference; and functional genomics with embryonic stem cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Austin, C.P. et al. The knockout mouse project. Nat. Genet. 36, 921–924 (2004).
Auwerx, J. et al. The European dimension for the mouse genome mutagenesis program. Nat. Genet. 36, 925–927 (2004).
Branda, C.S. & Dymecki, S.M. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6, 7–28 (2004).
van der Weyden, L., Adams, D.J. & Bradley, A. Tools for targeted manipulation of the mouse genome. Physiol. Genomics 11, 133–164 (2002).
Metzger, D. & Chambon, P. Site-and time-specific gene targeting in the mouse. Methods 24, 71–80 (2001).
von Melchner, H. & Stewart, A.F. Engineering of ES cell genomes with recombinase systems. in Handbook of Stem Cells, 609–623 (Elsevier, Burlington, Massachusetts, 2004).
Gardner, R.L. & Brook, F.A. Reflections on the biology of embryonic stem cells. Int. J. Dev. Biol. 41, 235–243 (1997).
Buehr, M. & Smith, A. Genesis of embryonic stem cells. Phil. Trans. R. Soc. Lond. B 358, 1397–1402 (2003).
Evans, M.J. & Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981).
Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634–7638 (1981).
Corcoran, L.M. & Metcalf, D. IL-5 and Rp105 signaling defects in B cells from commonly used 129 mouse substrains. J. Immunol. 163, 5836–5842 (1999).
Crawley, J.N. Behavioral phenotypes of inbred mouse strains: implications and recommendations for molecular studies. Psychopharmacology (Berl.) 132, 107–124 (1997).
Seong, E., Saunders, T.L., Stewart, C.L. & Burmeister, M. To knockout in 129 or in C57BL/6: that is the question. Trends Genet. 20, 59–62 (2004).
Tanaka, M., Hadjantonakis, A.K. & Nagy, A. Aggregation chimeras. Combining ES cells, diploid and tetraploid embryos. Methods Mol. Biol. 158, 135–154 (2001).
Eggan, K. & Jaenisch, R. Differentiation of F1 embryonic stem cells into viable male and female mice by tetraploid embryo complementation. Methods Enzymol. 365, 25–39 (2003).
Kunath, T. et al. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat. Biotechnol. 21, 559–561 (2003).
Seibler, J. et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 31, e12 (2003).
O'Gorman, S., Dagenais, N.A., Qian, M. & Marchuk, Y. Protamine–Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 94, 14602–14607 (1997).
Liu, P., Jenkins, N.A. & Copeland, N.G. Efficient Cre-loxP-induced mitotic recombination in mouse embryonic stem cells. Nat. Genet. 30, 66–72 (2002).
Belteki, G., Gertsenstein, M., Ow, D.W. & Nagy, A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing φC31 integrase. Nat. Biotechnol. 21, 321–324 (2003).
te Riele, H., Maandag, E.R. & Berns, A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc. Natl. Acad. Sci. USA 89, 5128–5132 (1992).
Zhang, Y., Buchholz, F., Muyrers, J.P. & Stewart, A.F. A new logic for DNA engineering using recombination in E. coli. Nat. Genet. 20, 123–128 (1998).
Copeland, N.G., Jenkins, N.A. & Court, D.L. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2, 769–779 (2001).
Zhang, Y., Muyrers, J.P., Testa, G. & Stewart, A.F. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18, 1314–1317 (2000).
Thomas, K.R., Deng, C. & Capecchi, M.R. High-fidelity gene targeting in embryonic stem cells by using sequence replacement vectors. Mol. Cell. Biol. 12, 2919–2923 (1992).
Hasty, P., Rivera-Perez, J. & Bradley, A. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell. Biol. 11, 5586–5591 (1991).
Valenzuela, D.M. et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol. 21, 652–659 (2003).
Yang, Y. & Seed, B. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat. Biotechnol. 21, 447–451 (2003).
Moens, C.B., Auerbach, A.B., Conlon, R.A., Joyner, A.L. & Rossant, J.A. Targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes Dev. 6, 691–704 (1992).
Skarnes, W.C. et al. The International Gene Trap Consortium. Nat. Genet. 36, 543–544 (2004).
Rodriguez, C.I. et al. High efficiency FLPe deleter mice provide a complement to Cre-loxP for in vivo genetic engineering. Nat. Genet. 25, 139–140 (2000).
Schaft, J., Ashery-Padan, R., van der Hoeven, F., Gruss, P. & Stewart, A.F. Efficient FLP recombination in mouse ES cells and oocytes. Genesis 31, 6–10 (2001).
Gu, H., Zou, Y.R. & Rajewsky, K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73, 1155–1164 (1993).
Ringrose, L., Chabanis, S., Angrand, P.-O., Woodroofe, C. & Stewart, A.F. Quantitative comparison of DNA looping in vitro and in vivo: Chromatin increases effective DNA flexibility at short distances. EMBO J. 18, 6630–6641 (1999).
Schnutgen, F. et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. USA 102, 7221–7226 (2005).
Albert, H., Dale, E.C., Lee, E. & Ow, D.W. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7, 649–659 (1995).
Schnutgen, F. et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat. Biotechnol. 21, 562–565 (2003).
Hansen, J. et al. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. USA 100, 9918–9922 (2003).
Zambrowicz B.P., et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl. Acad. Sci. USA 100, 14019–14114 (2003).
Keller, G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 19, 1129–1155 (2005).
Wobus, A.M. & Boheler, K.R. Embryonic stem cells: prospects for developmental biology and cell therapy. Physiol. Rev. 85, 635–678 (2005).
Testa, G. et al. A reliable expression reporter cassette for multipurpose, knock-out/conditional mouse alleles. Genesis 38, 151–158 (2004).
Adams, D.J. et al. Mutagenic insertion and chromosome engineering resource (MICER). Nat. Genet. 36, 867–871 (2004).
Lefebvre, L., Dionne, N., Karaskova, J., Squire, J.A. & Nagy, A. Selection for transgene homozygosity in embryonic stem cells results in extensive loss of heterozygosity. Nat. Genet. 27, 257–258 (2001).
Meister, G. & Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349 (2004).
Kittler, R. et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432, 1036–1040 (2004).
Berns, K. et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428, 431–437 (2004).
Seibler, J. et al. Single copy shRNA configuration for ubiquitous gene knockdown in mice. Nucleic Acids Res. 33, e67 (2005).
Ventura, A. et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. USA 101, 10380–10385 (2004).
Wiznerowicz, M. & Trono, D. Conditional suppression of cellular genes: lentivirus vector–mediated drug-inducible RNA interference. J. Virol. 77, 8957–8961 (2003).
Schmidt, E.E., Taylor, D.S., Prigge, J.R., Barnett, S. & Capecchi, M.R. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl. Acad. Sci. USA 97, 13702–13707 (2000).
Loonstra, A. et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA 98, 9209–9214 (2001).
Logie, C. & Stewart, A.F. Ligand-regulated site-specific recombination. Proc. Natl. Acad. Sci. USA 92, 5940–5944 (1995).
Forster, A. et al. Engineering de novo reciprocal chromosomal translocations associated with Mll to replicate primary events of human cancer. Cancer Cell 3, 449–458 (2003).
Herault, Y., Rassoulzadegan, M., Cuzin, F. & Duboule, D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE). Nat. Genet. 20, 381–384 (1998).
Spitz, F., Herkenne, C., Morris, M.A. & Duboule, D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat. Genet. 37, 889–893 (2005).
Zong, H., Espinosa, J.S., Su, H.H., Muzumdar, M.D. & Luo, L. Mosaic analysis with double markers in mice. Cell 121, 479–492 (2005).
Zheng, B. et al. Engineering a mouse balancer chromosome. Nat. Genet. 22, 375–378 (1999).
Baer, A. & Bode, J. Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr. Opin. Biotechnol. 12, 473–478 (2001).
Luo, J.L. et al. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene 20, 320–328 (2001).
Testa, G. et al. Engineering the mouse genome with bacterial artificial chromosomes to create multi-purpose alleles. Nat. Biotechnol. 21, 443–447 (2003).
Oram, M., Szczelkun, M.D. & Halford, S.E. Recombination. Pieces of the site-specific recombination puzzle. Curr. Biol. 5, 1106–1109 (1995).
Smith, M.C. & Thorpe, H.M. Diversity in the serine recombinases. Mol. Microbiol. 44, 299–307 (2002).
Olivares, E.C. et al. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat. Biotechnol. 20, 1124–1128 (2002).
Sauer, B. & McDermott, J. DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res. 32, 6086–6095 (2004).
Chen, Y. et al. Genotype-based screen for ENU-induced mutations in mouse embryonic stem cells. Nat. Genet. 24, 314–317 (2000).
Greber, B., Lehrach, H. & Himmelbauer, H. Mouse splice mutant generation from ENU-treated ES cells—a gene-driven approach. Genomics 85, 557–562 (2005).
Miskey, C., Izsvak, Z., Kawakami, K. & Ivics, Z. DNA transposons in vertebrate functional genomics. Cell. Mol. Life Sci. 62, 629–641 (2005).
Largaespada, D.A. Generating and manipulating transgenic animals using transposable elements. Reprod. Biol. Endocrinol. 1, 80 (2003).
Drabek, D. et al. Transposition of the Drosophila hydei Minos transposon in the mouse germ line. Genomics 81, 108–111 (2003).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005).
Bestor, T.H. Transposons reanimated in mice. Cell 122, 322–325 (2005).
Dupuy, A.J., Akagi, K., Largaespada, D.A., Copeland, N.G. & Jenkins, N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226 (2005).
Churchill, G.A. et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36, 1133–1137 (2004).
Fraser, A.G. & Marcotte, E.M. A probabilistic view of gene function. Nat. Genet. 36, 559–564 (2004).
Acknowledgements
We thank H. von Melchner, B. Skarnes, A. Smith, G. Testa and G. Yancopoulos for discussions. This work was supported by funding from the Sixth Research Framework Programme of the European Union, Project FunGenES and the VW Program on Conditional Mutagenesis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
A.F.S. is a shareholder in GeneBridges GmbH (DNA Engineering) and a consultant to SAB member Artemis Pharmaceuticals GmbH (Mouse Engineering).
Rights and permissions
About this article
Cite this article
Glaser, S., Anastassiadis, K. & Stewart, A. Current issues in mouse genome engineering. Nat Genet 37, 1187–1193 (2005). https://doi.org/10.1038/ng1668
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1668
This article is cited by
-
Neurogenic timing of the inferior olive subdivisions is related to the olivocerebellar projection topography
Scientific Reports (2023)
-
CRAGE enables rapid activation of biosynthetic gene clusters in undomesticated bacteria
Nature Microbiology (2019)
-
Skeletal muscle-specific Cre recombinase expression, controlled by the human α-skeletal actin promoter, improves glucose tolerance in mice fed a high-fat diet
Diabetologia (2018)
-
Cancer-specific binary expression system activated in mice by bacteriophage HK022 Integrase
Scientific Reports (2016)
-
RAC-tagging: Recombineering And Cas9-assisted targeting for protein tagging and conditional analyses
Scientific Reports (2016)