Abstract
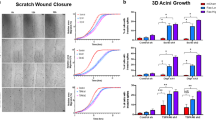
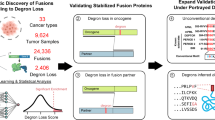
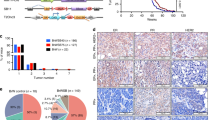
Truncation of Notch1 has been shown to cause a subtype of acute leukemia1, and activation of Notch4 has been associated with mammary and salivary gland carcinomas of mice2. Here we identify a new mechanism for disrupting Notch signaling in human tumorigenesis, characterized by altered function of a new ortholog of the Drosophila melanogaster Notch co-activator molecule Mastermind. We cloned the t(11;19) translocation that underlies the most common type of human malignant salivary gland tumor. This rearrangement fuses exon 1 from a novel gene of unknown function at 19p13, termed mucoepidermoid carcinoma translocated 1 (MECT1), with exons 2–5 of a novel member of the Mastermind-like gene family (MAML2) at 11q21 (ref. 3). Similar to D. melanogaster Mastermind and MAML1 (refs. 4,5), full-length MAML2 functioned as a CSL (CBF-1, suppressor of hairless and Lag-1)-dependent transcriptional co-activator for ligand-stimulated Notch. In contrast, MECT1–MAML2 activated transcription of the Notch target gene HES1 independently of both Notch ligand and CSL binding sites. MECT1–MAML2 induced foci formation in RK3E epithelial cells, confirming a biological effect for the fusion product. These data suggest a new mechanism to disrupt the function of a Notch co-activator in a common type of malignant salivary gland tumor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ellisen, L.W. et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661 (1991).
Jhappan, C. et al. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 6, 345–355 (1992).
Wu, L., Sun, T., Kobayashi, K., Gao, P. & Griffin, J.D. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol. Cell. Biol. 22, 7688–7700 (2002).
Artavanis-Tsakonas, S., Matsuno, K. & Fortini, M.E. Notch signaling. Science 268, 225–232 (1995).
Wu, L. et al. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26, 484–489 (2000).
Johansson, M. et al. Translocation 11;19 in a mucoepidermoid tumor of the lung. Cancer Genet. Cytogenet. 80, 85–86 (1995).
Horsman, D.E., Berean, K. & Durham, J.S. Translocation (11;19)(q21;p13.1) in mucoepidermoid carcinoma of salivary gland. Cancer Genet. Cytogenet. 80, 165–166 (1995).
El-Naggar, A.K., Lovell, M., Killary, A.M., Clayman, G.L. & Batsakis, J.G. A mucoepidermoid carcinoma of minor salivary gland with t(11;19)(q21;p13.1) as the only karyotypic abnormality. Cancer Genet. Cytogenet. 87, 29–33 (1996).
Dahlenfors, R., Wedell, B., Rundrantz, H. & Mark, J. Translocation(11;19)(q14–21;p12) in a parotid mucoepidermoid carcinoma of a child. Cancer Genet. Cytogenet. 79, 188 (1995).
Dahlenfors, R., Lindahl, L. & Mark, J. A fourth minor salivary gland mucoepidermoid carcinoma with 11q14–21 and 19p12 rearrangements. Hereditas 120, 287–288 (1994).
Mitelman, F. Recurrent chromosome aberrations in cancer. Mutat. Res. 462, 247–253 (2000).
Xu, T., Rebay, I., Fleming, R.J., Scottgale, T.N. & Artavanis-Tsakonas, S. The Notch locus and the genetic circuitry involved in early Drosophila neurogenesis. Genes Dev. 4, 464–475 (1990).
Jarriault, S. et al. Signalling downstream of activated mammalian Notch. Nature 377, 355–358 (1995).
Petcherski, A.G. & Kimble, J. Mastermind is a putative activator for Notch. Curr. Biol. 10, R471–R473 (2000).
Kojika, S. & Griffin, J.D. Notch receptors and hematopoiesis. Exp. Hematol. 29, 1041–1052 (2001).
Dumont, E. et al. Neoplastic transformation by Notch is independent of transcriptional activation by RBP-J signalling. Oncogene 19, 556–561 (2000).
Capobianco, A.J., Zagouras, P., Blaumueller, C.M., Artavanis-Tsakonas, S. & Bishop, J.M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 17, 6265–6273 (1997).
Petcherski, A.G. & Kimble, J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature 405, 364–368 (2000).
Wolfe, M.S. γ-Secretase inhibitors as molecular probes of presenilin function. J. Mol. Neurosci. 17, 199–204 (2001).
Hsieh, J.J. et al. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein–Barr virus EBNA2. Mol. Cell. Biol. 16, 952–959 (1996).
Wallberg, A.E., Pedersen, K., Lendahl, U. & Roeder, R.G. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by Notch intracellular domains in vitro. Mol. Cell. Biol. 22, 7812–7819 (2002).
Fryer, C.J., Lamar, E., Turbachova, I., Kintner, C. & Jones, K.A. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16, 1397–1411 (2002).
Tonon, G. et al. Spectral karyotyping combined with locus-specific FISH simultaneously defines genes and chromosomes involved in chromosomal translocations. Genes Chromosom. Cancer 27, 418–423 (2000).
Greenberg, R.A. et al. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 18, 1219–1226 (1999).
Tang, H.Y. et al. Constitutive expression of the cyclin-dependent kinase inhibitor p21 is transcriptionally regulated by the tumor-suppressor protein p53. J. Biol. Chem. 273, 29156–29163 (1998).
Kwon, T.K., Nagel, J.E., Buchholz, M.A. & Nordin, A.A. Characterization of the murine cyclin-dependent kinase inhibitor gene p27Kip1. Gene 180, 113–120 (1996).
Beatus, P., Lundkvist, J., Oberg, C. & Lendahl, U. The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development 126, 3925–3935 (1999).
Bessho, Y., Miyoshi, G., Sakata, R. & Kageyama, R. Hes7: a bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 6, 175–185 (2001).
Winter, E., Yamamoto, F., Almoguera, C. & Perucho, M. A method to detect and characterize point mutations in transcribed genes: amplification and overexpression of the mutant c-Ki-ras allele in human tumor cells. Proc. Natl. Acad. Sci. USA 82, 7575–7579 (1985).
Ruppert, J.M., Vogelstein, B. & Kinzler, K.W. The zinc-finger protein GLI transforms primary cells in cooperation with adenovirus E1A. Mol. Cell. Biol. 11, 1724–1728 (1991).
Acknowledgements
We are grateful to M. Kuehl, P. Aplan and T. Ried for helpful suggestions; P. Gao and J. Liu for their help; B. Baum for providing the HSY cells; M. Wolfe for providing the γ-secretase inhibitor; and A. Capobianco for advice on the RK3E assay. We acknowledge partial support by an American Society of Hematology Scholar Award and a General Motors Cancer Research Scholar Award (L.W.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
A patent application has been filed (by F.J.K. and G.T.) for the molecular diagnosis of mucoepidermoid carcinoma.
Supplementary information
Rights and permissions
About this article
Cite this article
Tonon, G., Modi, S., Wu, L. et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet 33, 208–213 (2003). https://doi.org/10.1038/ng1083
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1083
This article is cited by
-
The evolving role of molecular pathology in the diagnosis of salivary gland tumours with potential pitfalls
European Archives of Oto-Rhino-Laryngology (2022)
-
Mucoepidermoid carcinoma of the head and neck: CRTC1/3 MAML 2 translocation and its prognosticators
European Archives of Oto-Rhino-Laryngology (2022)
-
Whole-exome sequencing reveals the etiology of the rare primary hepatic mucoepidermoid carcinoma
Diagnostic Pathology (2021)
-
Targeting Notch and EGFR signaling in human mucoepidermoid carcinoma
Signal Transduction and Targeted Therapy (2021)
-
Does Nodal Metastasis and Perineural Invasion Affect Local Control in Hyalinizing Clear Cell Carcinoma of the Oral Cavity? A Case Report with Long Term Follow-Up
Head and Neck Pathology (2021)