Abstract
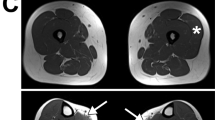
Hereditary inclusion body myopathy (HIBM; OMIM 600737) is a unique group of neuromuscular disorders characterized by adult onset, slowly progressive distal and proximal weakness and a typical muscle pathology including rimmed vacuoles and filamentous inclusions1. The autosomal recessive form described in Jews of Persian descent2 is the HIBM prototype. This myopathy affects mainly leg muscles, but with an unusual distribution that spares the quadriceps3. This particular pattern of weakness distribution, termed quadriceps-sparing myopathy (QSM), was later found in Jews originating from other Middle Eastern countries as well as in non-Jews4. We previously localized the gene causing HIBM in Middle Eastern Jews on chromosome 9p12–13 (ref. 5) within a genomic interval of about 700 kb (ref. 6). Haplotype analysis around the HIBM gene region of 104 affected people from 47 Middle Eastern families indicates one unique ancestral founder chromosome in this community. By contrast, single non-Jewish families from India, Georgia (USA) and the Bahamas, with QSM and linkage to the same 9p12–13 region, show three distinct haplotypes. After excluding other potential candidate genes, we eventually identified mutations in the UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase (GNE) gene in the HIBM families: all patients from Middle Eastern descent shared a single homozygous missense mutation, whereas distinct compound heterozygotes were identified in affected individuals of families of other ethnic origins. Our findings indicate that GNE is the gene responsible for recessive HIBM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Griggs, R.C. et al. Inclusion body myositis and myopathies. Ann. Neurol. 38, 705–713 (1995).
Sadeh, M. & Argov, Z. Hereditary inclusion body myopathy. in Inclusion Body Myositis and Myopathies: Jews of Persian Origin: Clinical and Laboratory Data. (eds. Askanas, V., Engel, W.K. & Serratrice, G.) 191–199 (Cambridge Press, Cambridge, 1997).
Argov, Z. & Yarom, R. “Rimmed vacuole myopathy” sparing the quadriceps. A unique disorder in Iranian Jews. J. Neurol. Sci. 64, 33–43 (1984).
Argov, Z. & Mitrani-Rosenbaum, S. Hereditary inclusion body myopathy (H-IBM) with quadriceps sparing: epidemiology and genetics. in Inclusion Body Myositis and Myopathies; (eds. Askanas, V., Engel, W.K. & Serratrice, G.) 200–210 (Cambridge Press, Cambridge, 1997).
Eisenberg, I. et al. Fine structure mapping of the hereditary inclusion body myopathy locus. Genomics 55, 43–48 (1999).
Eisenberg, I. et al. Physical and transcriptional map of the hereditary inclusion body myopathy locus on chromosome 9p12-p13. Eur. J. Hum. Genet. 9, 501–509 (2001).
Ponnambalam, S. et al. Chromosomal location and some structural features of human clathrin light-chain genes (CLTA and CLTB). Genomics 24, 440–444 (1994).
Takahashi, C. et al. Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc. Natl. Acad. Sci. USA 95, 13221–13226 (1998).
Hinderlich, S., Stasche, R., Zeitler, R. & Reutter, W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J. Biol. Chem. 272, 24313–24318 (1997).
Seppala, R., Lehto, V.P. & Gahl, W.A. Mutations in the human UDP-N-acetylglucosamine 2-epimerase gene define the disease sialuria and the allosteric site of the enzyme. Am. J. Hum. Genet. 64, 1563–1569 (1999).
Stasche, R. et al. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Molecular cloning and functional expression of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J. Biol. Chem. 272, 24319–24324 (1997).
Effertz, K., Hinderlich, S. & Reutter, W. Selective loss of either the epimerase or kinase activity of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase due to site-directed mutagenesis based on sequence alignments. J. Biol. Chem. 274, 28771–28778 (1999).
Salzberg, S.L., White, O., Peterson, J. & Eisen, J.A. Microbial genes in the human genome: lateral transfer or gene loss? Science 292, 1903–1906 (2001).
Keppler, O.T. et al. UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science 284, 1372–1376 (1999).
International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Claverie, J.M. Gene number. What if there are only 30,000 human genes? Science 291, 1255–1257 (2001).
Nonaka, I., Sunohara, N., Ishiura, S. & Satayoshi, E. Familial distal myopathy with rimmed vacuole and lamellar (myeloid) body formation. J. Neurol. Sci. 51,141–155 (1981).
Ikeuchi, T. et al. Gene locus for autosomal recessive distal myopathy with rimmed vacuoles maps to chromosome 9. Ann. Neurol. 41, 432–437 (1997).
Askanas, V. & Engel, W.K. Sporadic inclusion-body myositis and hereditary inclusion-body myopathies: current concepts of diagnosis and pathogenesis. Curr. Opin. Rheumatol. 10, 530–542 (1998).
Argov, Z., Eisenberg, I. & Mitrani-Rosenbaum, S. Genetics of inclusion body myopathies. Curr. Opin. Rheumatol. 10, 543–547 (1998).
Mitrani-Rosenbaum, S. et al. Hereditary inclusion body myopathy maps to chromosome 9p-q1. Hum. Mol. Genet. 5, 159–163 (1996).
Humbel, R. & Collart, M. Oligosaccharides in urine of patients with glycoprotein storage diseases. I. Rapid detection by thin-layer chromatography. Clin. Chim. Acta. 60, 143–145 (1975).
Little, S. Amplification-refractory mutation system (ARMS) analysis of point mutations. in Current Protocols in Human Genetics (eds. Dracopoli, N.C. et al.) 9.8.1–9.8.2 (Wiley, New York, 1995).
Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Higgins, D.G., Thompson, J.D. & Gibson, T.J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266, 383–402 (1996).
Acknowledgements
We are grateful to all of the family members who made these studies possible, and we extend our appreciation to S. Nazarian for her extensive efforts and our special thanks to M. Banayan for his endless and warm support. We also thank M. Korner, A.Tuvy, M. Mordechaishvili and all the staff from The Laboratory of DNA Analysis for skillful assistance in sequencing, A. Sanilevich for oligonucleotide synthesis, M.E. Ahearn for technical help, M. Zeigler for sialic acid measurements, D. Darvish and H. Raz for their involvement in blood collection, and T. Levi for her continuous support. This study was supported by Hadasit (Medical Research Services Development Co., a subsidiary for R&D of Hadassah Medical Organization; S.M.-R., Z.A.); by a special donation from Hadassah Southern California– Persian Group Council, Haifa Metro Group, Malka Group, Haifa San Diego Group, Vanguard II, Healing Spirit and the ARM organization (S.M.-R., Z.A.); by a special donation in memory of N. Hollo-Bencze (Z.A.); by an Israel Ministry of Science grant to the National Laboratory for Genome Infrastructure, The Crown Human Genome Center (D.L.); by the Krupp foundation (D.L.); and by the Weizmann Institute Glasberg, Levy, N. Brunschwig and Levine funds (D.L.). T.P. is a recipient of a Kamea fellowship from the Israeli Ministry of Science and Ministry of Absorption.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Eisenberg, I., Avidan, N., Potikha, T. et al. The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat Genet 29, 83–87 (2001). https://doi.org/10.1038/ng718
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng718
This article is cited by
-
Efficacy confirmation study of aceneuramic acid administration for GNE myopathy in Japan
Orphanet Journal of Rare Diseases (2023)
-
Inclusion body myositis: from genetics to clinical trials
Journal of Neurology (2023)
-
Different electrophysiology patterns in GNE myopathy
Orphanet Journal of Rare Diseases (2022)
-
Multidimensional analyses of the pathomechanism caused by the non-catalytic GNE variant, c.620A>T, in patients with GNE myopathy
Scientific Reports (2022)
-
The role of amyloid β in the pathological mechanism of GNE myopathy
Neurological Sciences (2022)