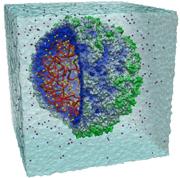
 The model shows one million atoms over 50 billionths of a second.Credit: University of Illinois / NCSA
The model shows one million atoms over 50 billionths of a second.Credit: University of Illinois / NCSAOne of the world's most powerful supercomputers has conjured a fleeting moment in the life of a virus. The researchers say the simulation is the first to capture a whole biological organism in such intricate molecular detail.
The simulation pushes today's computing power to the limit. But it is only a first step. In future researchers hope that bigger, longer simulations will reveal details about how viruses invade cells and cause disease.
Klaus Schulten at the University of Illinois, Urbana, and his colleagues built a computer model of the satellite tobacco mosaic virus, a tiny spherical package of RNA.
Their success depended on the latest version of a computer program called NAMD, which Schulten and his colleagues have built over the past decade to simulate biological molecules. The program allows the several hundred different processors within a supercomputer to work in parallel on the same problem.
Running on a machine at the National Center for Supercomputing Applications, Urbana, the program calculated how each of the million or so atoms in the virus and a surrounding drop of salt water was interacting with almost every other atom every femtosecond, or millionth of a billionth of a second.
The team managed to model the entire virus in action for 50 billionths of a second. Such a task would take a desktop computer around 35 years, says Schulten. "This is just a first glimpse," he says. "But it looks gorgeous."
In, out
The fleeting simulation, published in this month's Structure, reveals that although the virus looks symmetrical it pulses in and out asymmetrically, as if it were breathing1.
The model also shows that the virus coat collapses without its genetic material. This suggests that, when reproducing, the virus builds its coat around the genetic material rather than inserting the genetic material into a complete coat. "We saw something that is truly revolutionary," Schulten says.
Computer scientists have simulated viruses before, but they often had to limit themselves to one part of a virus and assume that the rest behaves in the same way.
ADVERTISEMENT
Other biologists study how proteins fold by harnessing many idle desktop computers. But Schulten says that this would not work for the virus simulation, because all the atoms must be modelled simultaneously.
Ultimately, computational biologists would like to simulate larger viruses such as influenza or the complex biological systems in a cell - and for longer periods, such as the thousandths of a second that it might take to observe proteins in a cell switch a gene off. These computer models should allow researchers to discover details about such processes that they may miss by observing a real virus.
But such simulations will not become possible until the next generation of supercomputers are built in the next five years, Schulten says.
Visit our buildsa_virus.html">newsblog to read and post comments about this story.
