Abstract
Small non-coding RNAs (sncRNAs) are potential vectors at the interface between genes and environment. We found that traumatic stress in early life altered mouse microRNA (miRNA) expression, and behavioral and metabolic responses in the progeny. Injection of sperm RNAs from traumatized males into fertilized wild-type oocytes reproduced the behavioral and metabolic alterations in the resulting offspring.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Manolio, T.A. et al. Nature 461, 747–753 (2009).
Qureshi, I.A. & Mehler, M.F. Nat. Rev. Neurosci. 13, 528–541 (2012).
Abe, M. & Bonini, N.M. Trends Cell Biol. 23, 30–36 (2013).
Rottiers, V. & Naar, A.M. Nat. Rev. Mol. Cell Biol. 13, 239–250 (2012).
Burton, N.O., Burkhart, K.B. & Kennedy, S. Proc. Natl. Acad. Sci. USA 108, 19683–19688 (2011).
Gu, S.G. et al. Nat. Genet. 44, 157–164 (2012).
Liu, W.M. et al. Proc. Natl. Acad. Sci. USA 109, 490–494 (2012).
Rassoulzadegan, M. et al. Nature 441, 469–474 (2006).
Kawano, M., Kawaji, H., Grandjean, V., Kiani, J. & Rassoulzadegan, M. PLoS ONE 7, e44542 (2012).
Krawetz, S.A. et al. Hum. Reprod. 26, 3401–3412 (2011).
Pena, F.J. et al. Reprod. Domest. Anim. 44, 345–349 (2009).
Franklin, T.B., Linder, N., Russig, H., Thony, B. & Mansuy, I.M. PLoS ONE 6, e21842 (2011).
Franklin, T.B. et al. Biol. Psychiatry 68, 408–415 (2010).
Weiss, I.C., Franklin, T.B., Vizi, S. & Mansuy, I.M. Front. Behav. Neurosci. 5, 3 (2011).
Rose, A.J. & Herzig, S. Mol. Cell. Endocrinol. 380, 65–78 (2013).
Drake, A.J. & Seckl, J.R. Pediatr. Endocrinol. Rev. 9, 566–578 (2011).
Sharma, A. Prog. Biophys. Mol. Biol. 113, 439–446 (2013).
El Ouaamari, A. et al. Diabetes 57, 2708–2717 (2008).
Zhang, N. et al. J. Biol. Chem. 288, 10361–10373 (2013).
Maguschak, K.A. & Ressler, K.J. J. Neuroimmune Pharmacol. 7, 763–773 (2012).
Tweedie-Cullen, R.Y., Reck, J.M. & Mansuy, I.M. J. Proteome Res. 8, 4966–4982 (2009).
Maragkakis, M. et al. BMC Bioinformatics 10, 295 (2009).
Hogan, B.C.F. & Lacy, L. Manipulating the Mouse Embryo: A Laboratory Manual, 2nd edn. (Cold Spring Harbor Laboratory, 1994).
Koshibu, K. et al. Protein phosphatase 1 regulates the histone code for long-term memory. J. Neurosci. 29, 13079–13089 (2009).
Sai Lakshmi, S. & Agrawal, S. Nucleic Acids Res. 36, D173–D177 (2008).
Li, H. & Durbin, R. Bioinformatics 25, 1754–1760 (2009).
Johnson, G.D. et al. Mol. Hum. Reprod. 17, 721–726 (2011).
Kozomara, A. & Griffiths-Jones, S. Nucleic Acids Res. 39, D152–D157 (2011).
Anders, S. & Huber, W. Genome Biol. 11, R106 (2010).
Betel, D., Sheridan, R., Marks, D.S. & Sander, C. PLoS Comput. Biol. 3, e222 (2007).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Genome Biol. 10, R25 (2009).
Acknowledgements
We thank M. Rassoulzadegan and V. Grandjean for help with the sperm purification, F. Manuella and H. Hörster for assistance with the MSUS paradigm, H. Welzl for help with behavior, G. Vernaz for help with western blotting, R. Tweedie-Cullen and P. Nanni for help with mass spectrometry, A. Patrignani for advice on DNA and RNA quality assessment, and A. Chen and A. Brunner for constructive discussions. This work was supported by the Austrian Academy of Sciences, the University of Zürich, the Swiss Federal Institute of Technology, Roche, the Swiss National Science Foundation, and The National Center of Competence in Research “Neural Plasticity and Repair”. P.S. was supported by a Gonville and Caius College fellowship.
Author information
Authors and Affiliations
Contributions
K.G. carried out all of the RT-qPCR, behavioral tests, metabolic measurements, and sperm RNA preparation for sequencing libraries and for RNA injection into fertilized oocytes, and part of the sequencing analyses. A.J. performed western blots and cell culture experiments and assisted with metabolic measurements. J.B. carried out the MSUS procedures and produced MSUS mice. J.P. and P.S. performed most RNA sequencing analyses. P.P. carried out the RNA injection experiments. E.M. and L.F. helped design the RNA sequencing analysis. K.G. and I.M.M. designed the study, interpreted the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Size and integrity profile of sperm RNAs used for deep sequencing and injected into fertilized oocytes.
Representative bioanalyser electropherograms show fluorescence intensity (fluorescence unit, FU) over time (seconds) during the pulsing of an RNA sample through a separation microchannel. Small RNAs go through the microchannel faster than long RNAs and appear on the left of the x-axis (for instance, 25bp RNAs appear after about 23 seconds, 200bp RNAs after 28 seconds and 2kb RNAs after 44 seconds). GQF-15 in a) corresponds to a control sample and GQF-14 in b) to a MSUS sample (pooled RNA from 5 mice). The profiles indicate that both samples contain short RNAs but no apparent RNAs above 0,5-1kb. They also show no DNA contamination. This was confirmed by Q-bit analyses using a dsDNA HS assay (Life technologies 1, [DNA]<0,1ng/ul). No protein contamination was detectable by Q-bit assay (Life technologies Q33212, [protein]<1pg/ul) and was confirmed by mass spectrometry (MS).
Supplementary Figure 2 SncRNAs in adult sperm.
Mapping of 15–44bp sequencing reads to a) the mouse reference genome, b) ribosomal RNAs, c) other non-coding RNAs and repeat regions and d) mitochondrial DNA, with multiple (black) or unique (grey) hits (n=16 mice, pooled in 4 samples). % total reads represents the proportion of reads with a given size mapping to the mouse genome or selected sequences over the total number of same-size reads. (e) Heatmap showing miRNAs (>100 reads) in control libraries which are altered by MSUS in adult sperm (n=3 each pooled from 5 mice). The blue-to-yellow scale is the number of normalized reads of a given sample over the mean normalized reads of all control samples for each miRNA. Bioinformatic analyses were performed twice using two independent methods. Data are mean ± s.e.m.
Supplementary Figure 3 Illustration of short RNA reads in adult mouse sperm.
(a) aligning to the mouse genome, (b) mapping to mature miRNA sequences (allowing for overhanging 5' and 3' nucleotides) and (c) aligning to piRNA clusters. In (a), reads alignment shows peaks at the typical size of miRNAs (21-23bp) and piRNAs (26-31bp). In (b), mapping of 18-35bp reads (not mapping to the transcriptome) to annotated miRNAs shows a sharp peak at 22bp, the typical size of mature miRNAs. In (c), alignment of 18-35bp reads (not mapping to the transcriptome) to genomic regions annotated as piRNAs shows a peak at the typical size of piRNAs, starting with the nucleotide T indicative for piRNA identity. A concatenation of all reads detected in control libraries is shown. The size and first nucleotide are shown by position on the x-axis and color, respectively. The y-axis shows the percentage of reads relative to total reads for the combined libraries.
Supplementary Figure 4 Proportion of 15-30bp reads including or excluding a 16bp specific sequence.
The y-axis represents the percentage of reads relative to total reads of combined control libraries either a) including an abundant 16bp sequence corresponding to an annotated piRNA sequence or b) excluding this 16bp sequence. Exclusion of this sequence results in a loss of the apparent enrichment of the 16bp peak, suggesting that the peak is not an artefact. A concatenation of all reads detected in control libraries is shown.
Supplementary Figure 5 Experimental design for MSUS treatment and breeding.
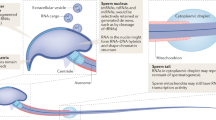
C57Bl/6J females (F0, left) were bred to C57Bl/6J males and their pups were subjected to MSUS from postnatal day (PND) 1 to 14 or raised in normal conditions (Control). Males from the F1 offspring were then bred to naïve C57Bl/6J females to obtain second-generation animals (F2) that were raised in normal conditions (no maternal separation or maternal stress). Animals from F1 and F2 generations were tested behaviorally then bred. Illustration: University of Zürich informatics services, MELS, Natasa Milosevic.
Supplementary Figure 6 Activity on an elevated plus maze.
Total distance covered by adult (a) F1 (controls n=8, MSUS n=17; t(23)=0.55),(b) F2 (controls n=30, MSUS n=30; t(53)=-1.06) and (c) RNA-injected (controls-RNAinj n=18, MSUS-RNAinj n=19, t(35)=0.18) animals. Data are mean ± s.e.m.
Supplementary Figure 7 Metabolic profile in F1 MSUS animals.
(a-c) Glucose level in blood a) at baseline and during a glucose tolerance test (GTT) after an acute restraint stress in non-fasted F1 mice (control, n=8; MSUS, n=8; F(1,22)=4.26) b) at baseline and during GTT in fasted F1 mice (control, n=8; MSUS, n=8; F(1,14)=0.01) c) at baseline and during an insulin tolerance test (ITT) in fasted F1 mice (control, n=8; MSUS, n=6; F(1,12)=5.38). (d) Body weight (control, n=10; MSUS, n=13; t(21)=1.82) and (e) caloric intake (control, n=4; MSUS, n=6; t(8)=-0.81) in F1 adult animals. Data are mean ± s.e.m. *p<0.05 group effect repeated measures ANOVA and t-test.
Supplementary Figure 8 Metabolic profile in F2 MSUS animals.
(a-b) Glucose level in blood (a) at baseline and during GTT in fasted F2 mice (control, n=8; MSUS, n=8; F(1,14)=4.71) and b) at baseline and during ITT in fasted F2 mice (control, n=7; MSUS, n=6; F(4,44)=3,38; 0 min: t(11)=-2.5, 15 min: t(11)=-0.15, 30 min: t(11)=2.76, 90 min: t(11)=-1.58). (c) Body weight (control, n=13; MSUS, n=11; t(21)=2.09) and (d) caloric intake (control, n=6; MSUS, n=6; t(6.52)=-2.44) in F2 adult animals. Data are mean ± s.e.m, *p<0,05 group effect repeated measures ANOVA and t-test.
Supplementary Figure 9 Effect of MSUS on piRNAs in adult sperm.
(a) Boxplot showing reads aligning to piRNA cluster 110 (on chromosome 13) per 1000 piRNAs reads in control and MSUS samples (negative binomial test p<0.1 after Bonferonni multiple test correction). (b) Log2 of the ratio of MSUS to control reads aligned to piRNA clusters on chromosome 13 showing that cluster 110 (in red) and two neighboring clusters (in black) are down-regulated in MSUS samples.
Supplementary Figure 10 MiRNA expression in hypothalamus and cortex in F1 mice.
Level of miRNAs expression in (a) hypothalamus (miR-375-3p: control, n=3; MSUS, n=4; t(5)=1.68; miR-375-5p: control, n=3; MSUS, n=4; t(5)=3.38; miR-200b-3p: control, n=3; MSUS, n=4; t(5)=-0.38; miR-672-5p: control, n=3; MSUS, n=4; t(6)=2.02; miR-466c-5p: control, n=3; MSUS, n=4; t(5)=1.98) and (b) cortex (miR-375-3p: control, n=4; MSUS, n=4; t(6)=0.81; miR-375-5p: control, n=4; MSUS, n=4; t(6)=-0.86; miR-200b-3p: control, n=4; MSUS, n=4; t(6)=-1.17; miR-672-5p: control, n=4; MSUS, n=4; t(6)=0.36; miR-466c-5p: control, n=4; MSUS, n=4; t(6)=0.53) in adult F1 control and MSUS males. Data are mean ± s.e.m, *p<0,05, #p<0,1.
Supplementary Figure 11 MiRNAs expression in the hippocampus of F3 animals.
Similar miRNA expression in F3 control and MSUS adult males (miR-375-3p: control, n=9; MSUS, n=9; t(16)=0.7; miR-375-5p: control, n=10; MSUS, n=9; t(17)=0.95; miR-200b-3p: control, n=9; MSUS, n=9; t(16)=0.51; miR-672-5p: control, n=10; MSUS, n=9; t(17)=-0.45; miR-466c-5p: control, n=10; MSUS, n=9; t(17)=0.58). Data are mean ± s.e.m.
Supplementary Figure 12 Analyses in MSUS-RNAinj males.
(a) Body weight of adult controls-RNAinj (n=8) and MSUS-RNAinj (n=9) animals (t(15)=1.9). (b) Level of miR-375-3p (Controls-RNAinj n=7, MSUS-RNAinj n=8; t(13)=.10) and miR-375-5p (controls-RNAinj n=7, MSUS-RNAinj n=7; t(12)=-2.3) in the adult hippocampus. Data are mean ± s.e.m, *p<0,05, #p<0,1.
Supplementary Figure 13 Performance of the offspring of RNAinj animals on a forced swim.
Time spent floating on the forced swim test in the offspring of Controls-RNAinj (n=19) and MSUS-RNAinj (n=12) animals (t(29)=-3.369). Data are mean ± s.e.m.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 and Supplementary Table 1 (PDF 3880 kb)
Rights and permissions
About this article
Cite this article
Gapp, K., Jawaid, A., Sarkies, P. et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 17, 667–669 (2014). https://doi.org/10.1038/nn.3695
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3695
This article is cited by
-
Dysfunction of DMT1 and miR-135b in the gut-testis axis in high-fat diet male mice
Genes & Nutrition (2024)
-
Semen proteome and transcriptome of the endangered black-footed ferret (Mustela nigripes) show association with the environment and fertility outcome
Scientific Reports (2024)
-
Stress Biomarkers Transferred Into the Female Reproductive Tract by Seminal Plasma Are Associated with ICSI Outcomes
Reproductive Sciences (2024)
-
Phthalates impact on the epigenetic factors contributed specifically by the father at fertilization
Epigenetics & Chromatin (2023)
-
Stress decreases spermatozoa quality and induces molecular alterations in zebrafish progeny
BMC Biology (2023)