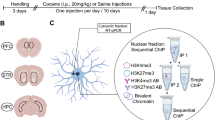
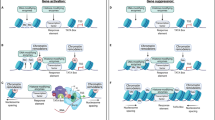
Abstract
Chronic exposure to drugs of abuse or stress regulates transcription factors, chromatin-modifying enzymes and histone post-translational modifications in discrete brain regions. Given the promiscuity of the enzymes involved, it has not yet been possible to obtain direct causal evidence to implicate the regulation of transcription and consequent behavioral plasticity by chromatin remodeling that occurs at a single gene. We investigated the mechanism linking chromatin dynamics to neurobiological phenomena by applying engineered transcription factors to selectively modify chromatin at a specific mouse gene in vivo. We found that histone methylation or acetylation at the Fosb locus in nucleus accumbens, a brain reward region, was sufficient to control drug- and stress-evoked transcriptional and behavioral responses via interactions with the endogenous transcriptional machinery. This approach allowed us to relate the epigenetic landscape at a given gene directly to regulation of its expression and to its subsequent effects on reward behavior.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Renthal, W. et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron 62, 335–348 (2009).
Kennedy, P.J. et al. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat. Neurosci. 16, 434–440 (2013).
Malvaez, M., Mhillaj, E., Matheos, D.P., Palmery, M. & Wood, M.A. CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. J. Neurosci. 31, 16941–16948 (2011).
Robison, A.J. & Nestler, E.J. Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 12, 623–637 (2011).
Feng, J. et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 15, R65 (2014).
Kumar, A. et al. SOM Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48, 303–314 (2005).
Kelz, M.B. et al. Expression of the transcription factor ΔFosB in the brain controls sensitivity to cocaine. Nature 401, 272–276 (1999).
Vialou, V. et al. ΔFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nat. Neurosci. 13, 745–752 (2010).
Robison, A.J. et al. Behavioral and structural responses to chronic cocaine require a feedforward loop involving ΔFosB and calcium/calmodulin-dependent protein kinase II in the nucleus accumbens shell. J. Neurosci. 33, 4295–4307 (2013).
Maze, I. et al. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc. Natl. Acad. Sci. USA 108, 3035–3040 (2011).
Covington, H.E. et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 71, 656–670 (2011).
Maze, I. et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327, 213–216 (2010).
Sun, H. et al. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J. Neurosci. 32, 17454–17464 (2012).
Snowden, A.W., Gregory, P.D., Case, C.C. & Pabo, C.O. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr. Biol. 12, 2159–2166 (2002).
Sanjana, N.E. et al. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171–192 (2012).
Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013).
Mendenhall, E.M. et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 31, 1133–1136 (2013).
Laganiere, J. et al. An engineered zinc finger protein activator of the endogenous glial cell line-derived neurotrophic factor gene provides functional neuroprotection in a rat model of Parkinson's disease. J. Neurosci. 30, 16469–16474 (2010).
Beerli, R.R. & Barbas, C.F. Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 20, 135–141 (2002).
Gerritsen, M.E. et al. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 94, 2927–2932 (1997).
Covington, H.E. et al. Antidepressant actions of histone deacetylase inhibitors. J. Neurosci. 29, 11451–11460 (2009).
Malvaez, M., Mhillaj, E., Matheos, D.P., Palmery, M. & Wood, M.A. CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. J. Neurosci. 31, 16941–16948 (2011).
Kennedy, P.J. et al. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat. Neurosci. 16, 434–440 (2013).
Kumar, A. et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48, 303–314 (2005).
Peixoto, L. & Abel, T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 38, 62–76 (2013).
Rogge, G.A., Singh, H., Dang, R. & Wood, M.A. HDAC3 is a negative regulator of cocaine-context-associated memory formation. J. Neurosci. 33, 6623–6632 (2013).
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).
Garriga-Canut, M. et al. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc. Natl. Acad. Sci. USA 109, E3136–E3145 (2012).
Smith, A.E., Hurd, P.J., Bannister, A.J., Kouzarides, T. & Ford, K.G. Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J. Biol. Chem. 283, 9878–9885 (2008).
Fritsch, L. et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP and SETDB1 participate in a multimeric complex. Mol. Cell 37, 46–56 (2010).
Filion, G. J. & van Steensel, B. Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nat. Genet. 42, 4 (2010).
Vialou, V. et al. Serum response factor and cAMP response element binding protein are both required for cocaine induction of FosB. J. Neurosci. 32, 7577–7584 (2012).
Zhang, F. et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 29, 149–153 (2011).
Maniatis, T., Goodbourn, S. & Fischer, J.A. Regulation of inducible and tissue-specific gene expression. Science 236, 1237–1245 (1987).
Ernst, J. & Kellis, M. Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types. Genome Res. 23, 1142–1154 (2013).
Chaudhury, D. et al. Rapid regulation of depression-related behaviors by control of midbrain dopamine neurons. Nature 493, 532–536 (2013).
Smith, D.J. & Konarska, M.M. A critical assessment of the utility of protein-free splicing systems. RNA 15, 1–3 (2009).
Grimmer, M.R. et al. Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation. Nucleic Acids Res. 42, 10856–10868 (2014).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Hathaway, N.A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012).
Cedar, H. & Bergman, Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304 (2009).
Paxinos, G. & Franklin, K.B.J. The Mouse Brain in Stereotaxic Coordinates (Academic Press, 2012).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Shen, L. et al. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE 8, e65598 (2013).
Lobo, M.K. et al. ΔFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional and optogenetic stimuli. J. Neurosci. 33, 18381–18395 (2013).
Becker, A. et al. Glutaminyl cyclase-mediated toxicity of pyroglutamate-beta amyloid induces striatal neurodegeneration. BMC Neurosci. 14, 108 (2013).
US National Research Council Committee on Guidelines for the Use of Animals in Neuroscience and Behavioral Research. Guidelines for the Care and Use of Mammals in Neuroscience and Behavioral Research (National Academies Press (US), 2003).
Maze, I. et al. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc. Natl. Acad. Sci. USA 108, 3035–3040 (2011).
Allan, R.S. et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature 487, 249–253 (2012).
Jennings, J.H. et al. Distinct extended amygdala circuits for divergent motivational states. Nature 496, 224–228 (2013).
Golden, S.A. et al. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat. Med. 19, 337–344 (2013).
Acknowledgements
The authors wish to thank G. Stuber and R. Ung for their help in generating social interaction heat maps. This work was supported by grants from the National Institute on Drug Abuse (E.A.H. and E.J.N.), the National Institute of Mental Health and the Hope for Depression Research Foundation (E.J.N.).
Author information
Authors and Affiliations
Contributions
E.A.H. designed and executed the biochemical, molecular and behavioral experiments (viral packaging constructs, qRT-PCR expression analysis, chromatin immunoprecipitation (mouse and human), immunohistochemical preparation and behavioral assays). H.M.C. and H.S. generated viral packaging constructs, conducted expression analysis and prepared chromatin. C.J.P. and S.A.G. analyzed immunohistochemical data. N.S. and L.S. conducted genome-wide sequence analysis. J.F. and E.A.H. performed DNA methylation sequencing. S.A.G. and S.J.R. generated human chromatin. E.A.H., H.M.C., H.S., J.J.W., M.M.-R., D.F. and M.-H.H. performed stereotaxic mouse surgery. J.P.H. generated mouse locomotor heat maps. C.S.T. prepared human postmortem brain tissue. R.L.N. generated HSV viral vectors. H.S.Z. generated the G9a catalytic domain construct. S.K., M.A.G. and C.N. generated the ZFP constructs. F.Z. generated the TALE constructs. C.J.P., E.A.H. and H.M.C. analyzed the data conducted statistical analyses. E.A.H. and E.J.N. discussed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 A suite of Fosb-ZFPs bidirectionally regulate Fosb expression in vitro.
(a) FosB/ΔFosB mRNA was significantly induced by several FosB-ZFP-p65 and -G9a constructs expressed in N2a cells and harvested after 48 hours. FosB-ZFP-NFD constructs activated gene expression to a lesser extent or not at all. Data are normalized to mock transfected cells. Complete statistics are available in Supplementary Table 3. Student's unpaired t-test: *P<0.05, *P<0.10. Data are presented as mean ± s.e.m.
Supplementary Figure 2 HSV-ZFPs specifically infect neurons in NAc and inhibit basal Fosb expression.
(a) HSV-GFP specifically infects DARPP-32 positive neurons in the NAc. White arrows indicate double labeled cells. (b) NAc injection of HSV-FosB-ZFP35-G9a repressed expression of FosB/ΔFosB in HSV infected (GFP+) cells [t13=3.55, *P=0.033; n=7, 8] compared to HSV-FosB-ZFP35-G9a (72 hours post HSV injection). Data are presented as mean ± s.e.m.
Supplementary Figure 3 Regulation of Fosb expression and reward behavior by Fosb-TALEs and a suite of additional catalytic domains fused to Fosb-ZFP35.
(a) Locations of FosB-ZFP and -TALE binding relative to the FosB TSS. The location of functional SRF and CREB sites are shown. (b) FosB/ΔFosB mRNA expression in the NAc was significantly induced by HSV-FosB-TALE2-VP64 [FosB: t8=3.03, *P=0.016; ΔFosB: t8=6.40, *P=0.000; n=5], and -FosB-TALE3-VP64 [FosB: t8=2.79, *P=0.023; ΔFosB: t8=4.01, *P=0.004; n=5] compared to control virus. (c) The binding sites of the 6-finger ZFP35 and 17-RVD (repeat variable diresidue) TALE1 recognize the FosB promoter at overlapping sites approximately 250 bp upstream from the FosB TSS. (d) FosB-ZFP35-p65 [FosB: t4=5.91, *P=0.004; ΔFosB: t4=26.11, *P=0.000; n=3], -p65x2 [FosB: t4=2.30, *P=0.000; ΔFosB: t4=3.46, *P=0.026; n=3], -VP16 [FosB: t4=9.04, *P=0.001; ΔFosB: t4=7.45, *P=0.002; n=3], and -VP64 [FosB: t4=19.40, *P=0.001; ΔFosB: t4=7.45, *P=0.001; n=3] and FosB-TALE1-VP64 [FosB: t4=6.80, *P=0.002; ΔFosB: t4=15.17, *P=0.000; n=3] activate FosB/ΔFosB mRNA levels when expressed in Neuro2a cells and harvested after 48 hours. Data are normalized to mock transfected cells. (e) HSV-FosB-TALE1-VP64 in NAc sensitizes cocaine-induced hyperactivity over time. There is a significant interaction between day, cocaine treatment, and virus [F(3,32)=3.42, *P=0.029]. TALE1-VP64 sensitizes the effect of cocaine on locomotor activity [main effect of day among TALE1-VP64 [F(3,35)=9.92, *P=0.000, n=10], but not GFP control [F(3,36)=2.05, P=0.126, n=9]. HSV-GFP data are the same as in Fig. 4d-e. Heat maps show representative locomotor data within the chamber for mice over the course of repeated cocaine exposure. Data are presented as mean ± s.e.m.
Supplementary Figure 4 Differential regulation by Fosb-ZFPs in vitro and in vivo.
(a) Expression of FosB-ZFP35-G9a differs between N2a cells and NAc [t6=4.50, *P=0.004; n=3,5]. (b) Titration of the amount of FosB-ZFP35-G9a expression in N2a cells leads to a reduction in the amount of induced mRNA expression of ΔFosB but not FosB. Using a 2-tailed Pearson correlation, we found a significant correlation between μg of transfected DNA and mRNA fold change of ΔFosB [R12=0.83, *P=0.001] but not FosB [R12=.66, P=0.665]. Data are presented as mean ± s.e.m.
Supplementary information
Supplementary Figures and Tables
Supplementary Figures 1–5 and Supplementary Tables 1–3 (PDF 18467 kb)
Supplementary Methods Checklist
Reporting Checklist for Nature Neuroscience (PDF 122 kb)
Rights and permissions
About this article
Cite this article
Heller, E., Cates, H., Peña, C. et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat Neurosci 17, 1720–1727 (2014). https://doi.org/10.1038/nn.3871
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3871
This article is cited by
-
Psychostimulant-induced aberrant DNA methylation in an in vitro model of human peripheral blood mononuclear cells
Clinical Epigenetics (2022)
-
Targeted demethylation at ZNF154 promotor upregulates ZNF154 expression and inhibits the proliferation and migration of Esophageal Squamous Carcinoma cells
Oncogene (2022)
-
Key transcription factors mediating cocaine-induced plasticity in the nucleus accumbens
Molecular Psychiatry (2022)
-
Drug addiction: from bench to bedside
Translational Psychiatry (2021)
-
Cocaine induces paradigm-specific changes to the transcriptome within the ventral tegmental area
Neuropsychopharmacology (2021)