Abstract
Current vaccine strategies against the asexual blood stage of Plasmodium falciparum are mostly focused on well-studied merozoite antigens that induce immune responses after natural exposure, but have yet to induce robust protection in any clinical trial. Here we compare human-compatible viral-vectored vaccines targeting ten different blood-stage antigens. We show that the full-length P. falciparum reticulocyte-binding protein homologue 5 (PfRH5) is highly susceptible to cross-strain neutralizing vaccine-induced antibodies, out-performing all other antigens delivered by the same vaccine platform. We find that, despite being susceptible to antibody, PfRH5 is unlikely to be under substantial immune selection pressure; there is minimal acquisition of anti-PfRH5 IgG antibodies in malaria-exposed Kenyans. These data challenge the widespread beliefs that any merozoite antigen that is highly susceptible to immune attack would be subject to significant levels of antigenic polymorphism, and that erythrocyte invasion by P. falciparum is a degenerate process involving a series of parallel redundant pathways.
Similar content being viewed by others
Introduction
The leading malaria vaccine candidate, 'RTS,S', achieves partial efficacy by blocking Plasmodium falciparum infection in the liver1; however, no vaccine has yet managed to emulate naturally acquired immunity by protecting against the disease-causing blood-stage parasite in clinical trials2. Two P. falciparum blood-stage antigens, merozoite surface protein 1 (PfMSP1)3 and apical membrane antigen 1 (PfAMA1)4, have dominated blood-stage vaccine development, but seem to require high antibody concentrations to induce protection and suffer antigenic diversity rendering vaccine-induced antibodies strain-specific5,6,7,8. There has never been a systematic head-to-head comparison of these and other candidate antigens delivered using the same human-compatible vaccine platform. More broadly, malaria vaccine development has been hampered by the difficulty of expressing recombinant plasmodial proteins, and by the need for potentially reactogenic chemical adjuvants to induce high-titre antibody responses in humans2.
There is thus a pressing need for validation of novel and recently identified antigens using technologies that allow rapid translation into clinical trials. We have recently shown that viral-vectored vaccines (replication-deficient adenoviruses and poxviruses that express antigen in vivo) can induce antibody responses in mice9,10,11, rabbits12,13, rhesus macaques14 and humans15 that compare favourably with leading protein-adjuvant formulations, and have an acceptable tolerability profile.
In this study, we produced viral-vectored vaccines expressing ten different P. falciparum blood-stage antigens and conducted a head-to-head comparison of their ability to induce antibodies in rabbits that neutralized parasites in the widely used in vitro assay of growth inhibitory activity. We report data that suggests that P. falciparum reticulocyte-binding homologue 5 (PfRH5) is an attractive candidate for blood-stage vaccine development.
Results
Vaccine production and immunogenicity
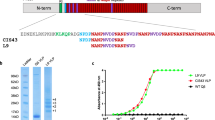
We developed viral-vectored vaccines expressing ten different P. falciparum (Pf) blood-stage antigens (Fig. 1a). These ten included the leading vaccine candidates PfMSP1 (refs 3, 13) and PfAMA1 (refs 4, 12); rhoptry-associated protein 3 (PfRAP3), which was selected on the basis that it is a close homologue of PfRAP2—an antigen that has demonstrated protection in a nonhuman primate P. falciparum challenge model16; merozoite surface protein 9 (PfMSP9), which was selected on the basis of evidence that it can induce growth inhibitory antibodies17; Pf38, which is a recently identified but untested potential vaccine antigen18,19; and five members of the erythrocyte binding-like and reticulocyte binding-like (or P. falciparum reticulocyte-binding homologue (PfRH)) proteins which have been implicated as important targets of the committed attachment process during red blood cell (RBC) invasion by P. falciparum merozoites20,21,22. All of the tested antigens are known to be expressed by P. falciparum 3D7 strain parasites with the exception of PfRH1 (refs 19, 23, 24, 25, 26, 27), and were based on the 3D7 strain sequence with the exception of P. falciparum erythrocyte binding antigen 175 (PfEBA175) F2 domain from the Camp strain28.
(a) Subcellular localization of vaccine-targeted merozoite antigens. (b) Indirect immunofluorescence images of P. falciparum schizonts stained with rabbit IgG (green) induced by 10 viral-vectored vaccines expressing malaria antigens, and negative control vectors lacking a malaria antigen. Nuclei are counterstained with 4,6-diamidino-2-phenylindole (blue). All sera were tested against 3D7 strain parasites, with the exception of anti-PfRH1 for which FVO strain parasites were used. All images to same scale as Giemsa-stained image (top left, on which scale bar indicates 5 μm). (c) Rabbit sera, taken two weeks after final immunization with PfEBA175, PfMSP9, Pf38, PfRH2 and PfRH5FL vaccines, contain IgG antibodies that recognize the corresponding recombinant proteins by ELISA at serum dilutions exceeding 1:10,000. Each point is the mean of two replicate wells for an individual rabbit. Line indicates group median.
Importantly, delivery of all ten antigens using the viral-vectored vaccine platform showed the antigens to be immunogenic in rabbits, inducing IgG antibodies that were able to recognize native malaria parasites as assessed by indirect immunofluorescence assay (IFA) (Fig. 1b), including cysteine-rich antigens such as Pf38 that can be difficult to express as recombinant proteins. Although the IFA data provide a sufficient confirmatory readout for induction of malaria antigen-specific IgG following a candidate vaccine-screening programme, we further confirmed antibody induction by vaccination when recombinant protein antigen was available by ELISA. The ELISA demonstrated antibody titres exceeding 10,000 for PfEBA175, PfRH2, PfRH5, PfMSP9 and Pf38 antigens (Fig. 1c); ELISA data demonstrating the immunogenicity of the PfMSP1 and PfAMA1 vaccines used here has been published elsewhere12,13.
3D7 parasite neutralization by vaccine-induced antibodies
We next tested the ability of vaccine-induced rabbit IgG to neutralize parasites in the widely used assay of growth inhibitory activity (GIA)7 against 3D7 strain P. falciparum parasites. This assay employs total purified IgG (of which only a fraction will be vaccine-induced and antigen-specific). IgG induced by vectors expressing the antigen PfRH5FL (full-length reticulocyte-binding protein homologue 5) was potently growth inhibitory (Fig. 2a). The potent inhibition shown by anti-PfRH5FL IgG contrasted with modest inhibition by antibodies induced by vectors expressing a PfRH5 fragment that, when previously tested as a bacterially expressed immunogen, had failed to show significant GIA in vitro when using intact human RBCs24. Another vaccination study utilizing a different bacterially expressed fragment of the PfRH5 antigen also reported a failure to show GIA in vitro29. The conformational accuracy of these bacterially expressed fragments is unknown; for now, the full-length PfRH5 immunogen seems to be required to induce functional growth inhibitory antibodies following immunization. Unlike any other reported antigen to date, anti-PfRH5FL IgG was more effective than antibodies to the leading antigens PfAMA1 and PfMSP1 when tested against vaccine-homologous 3D7 parasites (the strain used for controlled human malaria infection vaccine-efficacy trials30, and on which the vaccine antigen was based).
(a) GIA of rabbit IgG tested against 3D7 strain parasites at various concentrations. IgG from all rabbits immunized with each antigen was pooled, with the exception of PfAMA1 and PfRH5FL, for which GIA was performed with IgG purified from individual rabbits. Values are mean of two independent experiments, typically with three replicate wells. Error bars indicate inter-well s.d., with the exception of PfAMA1 and PfRH5FL, for which bars indicate inter-rabbit s.d. (b) Comparison of effects of PfRH5FL-induced IgG (blue), IgG induced by vaccination with 3D7-strain PfAMA1 (green), and IgG induced by vaccination with a bivalent PfAMA1 vaccine (red) in assays of GIA against parasite strains 3D7, FVO, Dd2, GB4, Camp and 7G8. Lines indicate dose-response curves fitted by nonlinear least squares regression as described in methods section. All assays were performed using IgG purified from pooled serum of two PfRH5FL-vaccinated rabbits and single rabbits vaccinated with each PfAMA1 vaccine (see Methods for further details). Values are mean of 2 independent experiments with 3 replicate wells, with the exception of assays against 7G8 and Camp strains and monovalent PfAMA1 versus FVO (for which results are mean of 3 wells in a single experiment). Error bars indicate inter-well SD.
Data generated, using a twofold dilution of total IgG, allows for a more accurate comparison of the effect of anti-PfRH5FL IgG with that of anti-PfAMA1 IgG. This shows that the effective concentration of total IgG from PfRH5FL-immunized rabbits, required to give 50% GIA (EC50) against 3D7 strain parasites, is twofold lower than that observed with PfAMA1 (Fig. 2b; Table 1). Given that exceptionally high levels of anti-PfAMA1 antibodies (and correspondingly high levels of GIA) do seem to be associated with protection in nonhuman primates and trend towards protective effects in human challenge6,31,32, such reductions in the antibody concentration required to achieve an effect could facilitate the achievement of a protective level of GIA in humans.
Neutralization of vaccine-heterologous parasites
We proceeded to test the anti-PfRH5FL-induced rabbit IgG in assays of GIA against vaccine heterologous parasite strains. Sequencing of the PfRH5 gene in 18 laboratory parasite strains has previously showed limited polymorphism (10 amino acid sites)33. We thus selected five vaccine-heterologous parasite strains (FVO, Dd2, GB4, Camp and 7G8) on the basis of their genetic divergence from 3D7's PfRH5 sequence, and their differing patterns of ligand–receptor invasion 'pathway' usage33,34. Together, these strains included amino acid changes at every polymorphic locus previously identified. Additionally, FVO, Dd2, Camp and GB4 parasites are known to vary substantially from 3D7 parasites in their ability to invade Aotus nancymaae erythrocytes, a phenotype believed to be associated with PfRH5 polymorphisms that may affect receptor recognition33. Of 18 laboratory strains for which the PfRH5 gene has been sequenced and Aotus RBC invasion efficiency quantified, FVO was most divergent from the 3D7 strain on which the vaccine was based33.
Strikingly, antibodies against PfRH5FL remained highly effective against all five extra parasite strains tested, in marked contrast to antibodies induced by both monovalent (3D7) and bivalent (3D7 and FVO) PfAMA1 vaccines (Fig. 2b). Using the same data, we estimated EC50 values for the GIA effect of anti-PfRH5FL and anti-PfAMA1 IgG against the various parasite strains (Table 1). The EC50 of anti-PfRH5FL was even lower against vaccine-heterologous parasite strains than against 3D7 parasites. Taken together with the higher EC50 values for anti-PfAMA1 against vaccine-heterologous parasites, this resulted in a >5-fold reduction in EC50 for anti-PfRH5FL relative to anti-PfAMA1 against all vaccine-heterologous parasite strains. The breadth of cross-strain parasite-neutralizing activity, induced by the PfRH5FL antigen, may thus represent an intriguing gap in P. falciparum's highly evolved immune evasion strategies and is, to our knowledge, unique to date among monovalent blood-stage vaccine candidates.
Our key observations were independently confirmed, with a notable degree of similarity, by the GIA Reference Center funded by the PATH malaria vaccine initiative (MVI) (Fig. 3), as recommended by other studies of invasion-ligand candidate vaccines20. Given the lack of PfRH1 expression by 3D7 parasites, we additionally tested our anti-PfRH1 IgG against FVO parasites, which do express this antigen27,35; no GIA was detectable at 10 mg ml−1 total IgG.
GIA performed at the PATH MVI funded GIA reference laboratory at NIH, USA against 3D7 strain parasites (a) and FVO parasites (b). Blue, PfRH5FL (3D7) and red, AMA1 biallelic (3D7 and FVO). Error bars indicate range of results for the two PfRH5FL-vaccinated rabbits. (c) Comparison of GIA results obtained in Oxford and at NIH. Each point represents the result obtained for a single rabbit at a single IgG concentration. Pearson r=0.90.
Assessment of naturally acquired PfRH5 antibody responses
We hypothesize that, despite being vulnerable to antibody, PfRH5 may not be under substantial immune selection pressure. Consistent with this, we found that malaria-experienced Kenyans have little or no antibody against PfRH5, in contrast to all other antigens we were able to test for which recombinant protein was available as an ELISA-coating antigen (Fig. 4a,b). Mammals may be tolerant to proteins with extensive primary amino acid homology with mammalian proteins, but this is not the case for PfRH5; a Protein BLAST search yielded no match with an E-value<1 between PfRH5 and any mouse or human protein; the closest match for both species was a SLIT-ROBO RhoGTPase, with limited similarity to residues 160–303 of PfRH5 (within this limited stretch, PfRH5 had 26% sequence identity with the human GTPase and 27% identity with the mouse homologue). Indeed, antibodies to PfRH5FL are readily inducible by vaccination of mice (geometric mean endpoint titre 85,000, range 35,000–216,000, n=5) and rabbits (Fig. 1c), confirming the antigen is not inherently non-immunogenic in mammals.
(a) Semi-quantitative comparison of levels of IgG (as indicated by optical density, OD 405 nm, in ELISA) against multiple antigens in 24 Kenyan adult sera relative to background in 6 malaria-naïve healthy UK adults. Boxes enclose interquartile range, central lines represents median, whiskers indicate 10th and 90th centile. Blue, Kenyan and red, UK. (b) Prevalence of antigen-specific IgG among 24 Kenyan adult sera (samples from 3A). The threshold for positivity was defined as the mean OD 405 nm plus 3 standard deviations of results for 6 malaria-naïve UK adults. (c) Age-related acquisition of IgG antibodies to blood-stage antigens, such as PfAMA1 is commonly observed in children in malaria-endemic areas. The data compare acquisition of anti-PfAMA1 and anti-PfRH5 IgG in 55–60 children from each of 5 age groups; acquisition of antigen-specific IgG is minimal for PfRH5. Boxes enclose interquartile range, central lines represents median, whiskers indicate 10th and 90th centile. Blue, AMA1 and red, RH5.
Discussion
The reason for the lack of anti-PfRH5 antibody in naturally exposed Kenyans is unclear. The facts that anti-PfRH5FL IgG induced in animals by these viral-vectored vaccines binds to both the ELISA antigen and native parasites by IFA (Fig. 1b), and that our ELISA coating antigen has been confirmed to bind to its host RBC receptor36, indicate that incorrect conformation of our PfRH5 ELISA coating antigen is highly unlikely to account for poor detection of anti-PfRH5 IgG in humans. PfRH5 messenger RNA has also been shown to be expressed in field isolates cultured ex-vivo both from The Gambia in West Africa37, and Kenya in East Africa38, suggesting this inherent lack of immune recognition is not due to the absence of PfRH5 expression by parasites circulating in endemic areas. Selective downregulation of PfRH5 in parasites infecting malaria-experienced individuals who control parasitemia (as suggested by Gomez-Escobar et al.37) might lead anti-PfRH5 IgG titres to be higher in children than in adults. However, further ELISA assays conducted using serum from Kenyan children (Fig. 4c) yielded results comparable to those observed in adults (Fig. 4a). The mechanism by which this crucial antigen escapes immune recognition in both adults and children thus requires further investigation.
The density of known polymorphisms in PfRH5 (10 out of 526 amino acids) is 3.3-fold lower than in the well-characterized 19 kDa Carboxy terminus of PfMSP1 (PfMSP119) (6 out of 96 amino acids)39, which in itself is widely regarded as a highly conserved vaccine target. Data from nonhuman primate models demonstrate that the amino acid polymorphism in PfMSP1 can result in strain-specific efficacy of vaccine-induced anti-PfMSP1 antibody responses5. There may thus be a contrast in the immunological effect of the polymorphism in PfMSP1 versus that in PfRH5. It is possible that non-immune mechanisms such as adaptation to host erythrocyte variability could drive the apparent excess of non-synonymous relative to synonymous mutations in PfRH5 (refs 33, 40, 41).
Attempts to delete the PfRH5 gene have failed repeatedly, suggesting it is essential for parasite viability24,33, and immunization against rodent malaria homologues of the PfRH family provides protection in a mouse challenge model42—a finding first reported in 1980, but not translated into a human-compatible vaccine formulation against P. falciparum until now. Complementing our data obtained using PfRH5FL as a vaccine target, we have recently identified an erythrocyte receptor for PfRH5 and demonstrated a critical role for this receptor–ligand interaction36. In contrast to the prevailing views that erythrocyte invasion by P. falciparum is a degenerate process mediated by multiple ligand–receptor pairs and that any merozoite antigen susceptible to antibody is highly polymorphic, PfRH5-mediated invasion seems to be both essential and vulnerable to blockade.
Multiple strands of evidence now suggest that PfRH5FL is the best candidate for blood-stage malaria vaccine development to emerge from the P. falciparum genome to date. The traditional immuno-epidemiological approach to antigen identification may have focussed vaccine-development efforts more on immunogenic but polymorphic 'decoy antigens' than 'Achilles' heels'43,44. Assaying parasite growth inhibition by vaccine-induced antibodies has, instead, directly demonstrated strain-transcending antibody effects obtained with an antigen that would not have seemed to be an important target of naturally acquired responses. In this study, anti-PfRH5FL IgG outperformed anti-PfAMA1 IgG against homologous 3D7 strain parasites when induced by the same vaccine delivery platform, and, importantly for clinical significance, also showed strain-transcending neutralization against other strains. Recently a 3D7 strain AMA1 protein-in-adjuvant vaccine has demonstrated significant strain-specific efficacy in a pre-specified secondary analysis of a Phase IIb study in Malian children8. This result further encourages the development of blood-stage vaccines, but highlights that antigenic polymorphism is a central problem for the field and the one currently shared by all leading blood-stage vaccine candidate antigens. Our findings reported here suggest that the PfRH5FL antigen offers a solution to this problem.
P. falciparum possesses over 5,000 genes. Although the genome sequence of P. falciparum was published in 2002 (ref. 45), PfMSP3 (first described in 1994 (ref. 46)) is the most recently identified antigen to have entered clinical trials as a candidate blood-stage vaccine2. Although genome sequence data has substantially advanced basic scientific knowledge, vaccinologists working on complex human pathogens such as P. falciparum have yet to fully exploit pathogen genome data to identify, comparatively assess and validate much needed novel antigens47. This study has highlighted PfRH5FL from a small candidate panel: continued and similar efforts with a larger panel of antigens should thus yield further promising pre-erythrocytic, blood-stage and sexual-stage malaria vaccine candidates as well as permit truly effective exploitation of the genomes of other challenging vaccine targets.
Methods
Viral vectors
P. falciparum antigen genes were codon-optimized for human expression and synthesized by GeneArt GmbH. Unless otherwise stated, antigens were based on the 3D7 strain coding sequence. PfEBA175, PfRH1, PfRH2, and PfRH4 constructs were based on fragments known to include erythrocyte-binding domains25,27,28,48. Remaining constructs encoded full-length proteins, omitting signal peptides and transmembrane domains where applicable. In some cases, predicted N-glycosylation sites were removed by N—Q or S—A substitution.
The following antigens were used (GenBank Nucleotide core accession codes are provided): PfEBA175, Genbank X52524 from Camp strain, amino acids 447–795, ELRE...RDDD, the 'F2' erythrocyte-binding region28; PfMSP9, NCBI XM_001350647.1, amino acids 25–719, KNDK...EESK, with substitutions N529Q, N580Q, N621Q; Pf38, NCBI XM_001351602.1, amino acids 23–327, VENK...REEI, with substitutions S166A, N294Q, S297A, N301Q; PfRAP3, NCBI XM_001351538.1, amino acids 23–399, NKCK...NIFK, with substitutions N50Q, N246Q; PfRH1, NCBI XM_002808591.1, amino acids 500–833, LQIV...LTN, with substitutions N685Q, N830Q; PfRH2, from the PfRH2a sequence NCBI XM_001350047.1, amino acids 2,030–2,531, ELRE...MLLN, a sequence shared between PfRH2a and PfRH2b and previously termed PfRH2A9 (ref. 48) (becomes 48); PfRH4, NCBI XM_001351509, amino acids 28–766, PSKE...MQNI and previously termed PfRH4.9 (ref. 25); PfRH5 fragment (PfRH5frag), NCBI XM_001351508.1, amino acids 191–359, NSIY...IRYH, as previously expressed in E.scherichia coli24; PfRH5 full-length (PfRH5FL), NCBI XM_001351508.1, amino acids 26–526, ENAI...PLTQ, with substitutions N38Q and N214Q.
Antigens were cloned into replication-deficient adenovirus human serotype 5 (AdHu5) and poxvirus (modified vaccinia virus Ankara, MVA) genomes downstream of a mammalian secretory signal (from bovine tissue plasminogen activator), and viruses prepared using previously described techniques for MVA9 and AdHu5 (ref. 49). AdHu5 was chosen as a model adenovirus without intellectual property restrictions and suitable for antigen screening. The immunogenicity and efficacy of AdHu5 vaccines can be readily matched by clinically applicable simian adenoviruses: we have recently shown that the immunogenicity of AdHu5 and various simian adenoviruses is comparable in animal studies12,13,50, and that a chimpanzee adenovirus serotype 63 (ChAd63) virus expressing PfMSP1 is highly immunogenic and safe in healthy adult volunteers as assessed in a Phase Ia clinical trial15.
AdHu5 viruses expressed antigen under the control of the intron-containing native cytomegalovirus immediate-early promoter, modified in the case of PfMSP9, Pf38, PfRAP3, PfRH1, PfRH4 and PfRH5FL expressing viruses to include tandem tetracycline operators in proximity to the TATA box by ligation of a fragment containing these into the SacI restriction site51,52. Such 'tet repressed' viruses were cultured on Trex293 cells (Invitrogen). The production of monovalent (3D7) and bivalent (3D7 and FVO) adenoviral- and MVA-vectored vaccines against PfMSP1 (previously termed 'PfM128') and PfAMA1 has been described elsewhere12,13,14. Viral-vectored vaccines that did not express malaria antigens were used as negative controls (an AdHu5 virus without an antigen insert, and an MVA virus expressing an influenza antigen)10,53. Adenoviruses were titred by immunostaining, carried out by infection of Trex293 cells with serial dilutions of virus. Forty-eight hours post-infection, cells were stained with anti-hexon antibody (Cambridge Bioscience) and detected with HRP-conjugated secondary antibody (Cambridge Bioscience) and ImmPact DAB reagent (Vector Labs).
Recombinant proteins
GST-tagged PfRH2A9 (referred to in the ELISA data as PfRH2) was produced, as described previously for the 3D7 ('ETSR') allele of the 19 kDa region of PfMSP1 (PfMSP119)13. The latter is referred to as PfMSP1 in the ELISA data. An amino-terminally His6-tagged PfRH4 fragment (referred to in the ELISA data as PfRH4, NCBI refseq XM_001351509 from 3D7 strain, amino acids 1,329–1,607, NHIK...NAYY, the most C-terminal region before the transmembrane domain, not overlapping with our vaccine antigen54) was produced by cloning the fragment into the pTrcHisC plasmid (Invitrogen), transforming Rosetta-strain E. coli, inducing expression and purifying protein, according to the instructions of the Qiagen Ni-NTA fast-start kit (Qiagen). Recombinant PfEBA175_F2 (referred to in the ELISA data as PfEBA175) and 3D7 strain PfAMA1 (Fig. 4a,b) were produced as previously described and were a kind gift from Dr Chetan E Chitnis (ICGEB, New Delhi, India)12,14,28.
PfAMA1 (used for the ELISA in Fig. 4c), PfMSP2, PfMSP9, Pf38 and PfRH5 proteins were produced as enzymatically monobiotinylated soluble recombinant proteins by transient transfection of HEK293E cells, as previously described36,55. Supernatants were extensively dialysed against PBS, expression levels normalized, and captured on streptavidin-coated microtitre plates for ELISAs. The PfRH5 protein produced in this manner was confirmed to bind to its erythrocyte receptor, basigin, by surface plasmon resonance36.
Animals and immunization regimes
Mouse and rabbit work was approved by the University of Oxford Animal Care and Ethical Review Committee (in its review of the application for Home Office project license PPL 30/2414), and performed in accordance with all applicable regulations.
Six-week-old Balb/c female mice were housed under specified-pathogen-free conditions, immunized with 1×108 infectious units AdHu5 PfRH5FL, and boosted with 1×107 plaque-forming units MVA PfRH5FL 122 days later. Sera were collected 14 days after the boost immunization.
Rabbit work was conducted by Agrobio, and Biogenes. Female New Zealand white rabbits (2–4 per group) housed under nonspecified-pathogen-free conditions were immunized with 7×107–4.5×108 infectious units of recombinant AdHu5 on day 0, 5×107–1×108 plaque-forming units MVA on day 56, and, in the case of the PfRH2 and PfEBA175 groups, received a third immunization on day 114 with 100 μg of either PfRH2 or PfEBA175 recombinant protein mixed with 20 μl (18 μg) Abisco adjuvant (ISCOM Matrix M)11,14,56. Serum was collected two weeks after final vaccination. Rabbits immunized with the same negative control vaccines and positive control vaccines (bivalent PfAMA1) were included in each study.
Indirect immunofluorescence assay
Parasite cultures containing mainly schizonts were smeared onto slides, fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X100 and quenched with 0.01% sodium borohydride. Slides were then blocked for 1 h in 10% goat serum, 1% BSA in PBS before the addition of purified rabbit IgG (see below). Bound IgG was detected with goat anti-rabbit IgG-Alexa 488 conjugate (Invitrogen). Nuclear DNA was counterstained with 4,6-diamidino-2-phenylindole. Slides were viewed under a Leica DMI3000 microscope. 3D7 strain parasites were used for all IFA except for PfRH1, for which the FVO strain was used, as this antigen is known to be poorly expressed in 3D7 (ref. 27).
Human serum samples
UK Adult sera (Fig. 4a,b) were obtained from pre-vaccination samples taken from healthy malaria-naïve adult volunteers enrolled in a Phase I/IIa malaria vaccine clinical trial with appropriate ethical approval (EudraCT number 2008-006804-46, OXREC-A reference 09/H0604/9)15.
Kenyan adult sera (Fig. 4a,b) were collected during adult cross-sectional surveys between 2006 and 2008 from the villages surrounding the Chonyi area in Kilifi, Kenya that experiences moderate malaria transmission with an EIR of 10–100 infective bites/person/year; these adults are considered to have substantial naturally acquired immunity as evidenced by the decline in clinical episodes of malaria with age57. Scientific and ethical approvals for the Kenyan serum samples were granted by the Kenya National Scientific and Research Ethics Committees, respectively, SSC No. 1131.
Kenyan Child sera (Fig. 4c) samples were obtained from a cohort that has been extensively used to analyse naturally acquired antibodies against leading blood-stage malaria vaccine candidate antigens58. The present work focused on children aged up to 10 years of age because they accounted for nearly 90% of all the malaria episodes and thus are in the process of actively acquiring immunity.
ELISA
To generate the rabbit ELISA data in Figure 1c, PfRH2, PfRH4 and PfEBA175 proteins (described above) were coated on Nunc Maxisorp plates at 100 ng per well, or wells of streptavidin-coated plates (Nunc Immobilizer) were saturated with biotinylated Pf38, PfMSP2, PfMSP9 or PfRH5 in PBS and left over-night. The next day, plates were washed 6× in PBS containing 0.05% Tween 20 (PBS/T). Plates were blocked with 10% skimmed milk in PBS/T for 1 h, dilution series of serum samples (threefold from 1:300) were added for 2 h, before the plates were washed again. Alkaline-phosphatase-conjugated goat anti-rabbit IgG (Sigma) was used for detection. Plates were washed again, and bound antibodies were detected by adding p-nitrophenylphosphate substrate (pNPP, Sigma) diluted in diethanolamine buffer (Fisher Scientific). OD 405 nm was read 20 min after addition of substrate, using an ELx800 microplate reader (BioTek). End-point titres were taken as the x-axis intercept of the dilution curve at an absorbance value three standard deviations (s.d.) greater than the OD405 for naïve rabbit serum. Mouse ELISA was conducted using an identical method, with the exception of the use of alkaline-phosphatase-conjugated goat anti-mouse IgG (Sigma) for detection.
To generate the human ELISA data in Figure 4a,b, GST–PfMSP119 (ETSR 3D7/Mad20) or 3D7 AMA1 (described earlier) protein was coated onto 96-well Nunc Immuno Maxisorp plates at a concentration of 2 μg ml−1 in PBS and left overnight. The next day, plates were washed 6x in PBS containing 0.05% Tween 20 (PBS/T) and blocked for 1 h with Casein block solution (Pierce). Plates were washed again, and, then, a standard, test sera, internal control and blank samples all diluted in Casein block solution were added to each plate for 2 h according to published methodology15,59. The standard was prepared from adult Kenyan immune serum and was serially diluted on every plate to make a standard curve. Plates were washed again, followed by addition for 1 h of alkaline phosphatase-conjugated goat anti-human IgG (γ-chain) (Sigma) diluted 1:1,000 in Casein block solution. Plates were washed and developed as above. The ELISA unit value of the standard was assigned as the reciprocal of the dilution giving an OD405 of 1.0 in the standardized assay. The OD405 of individual test samples was converted into ELISA units by using the standard curve and Gen5 ELISA software v1.10 (BioTek). All sera tested against the GST control protein were less than the minimal detection level of the assay.
Alternatively, PfRH2, PfRH4 and PfEBA175 proteins were coated on Nunc Maxisorp plates at 100 ng per well, or wells of streptavidin-coated plates (Nunc Immobilizer) were saturated with biotinylated Pf38, PfMSP2, PfMSP9 or PfRH5 in PBS and left overnight. All test sera were diluted 1:300 in Casein block solution. Washing, blocking and detection were as for PfMSP1 and PfAMA1. OD 405 nm was measured 35 min after substrate addition, using an ELx800 microplate reader (BioTek).
To generate the human ELISA data in Figure 4c, streptavidin-coated plates (NUNC Immobilizer Streptavidin, Thermo Fisher Scientific) were washed 3 times with PBS-Tween (PBS-0.1% Tween 20). Individual wells were coated with 100 μl of antigen and incubated for 45 min at room temperature (RT). Plates were then washed 4 times with Hepes-buffered saline-Tween (HBS-Tween: 0.14 M NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 0.1% Tween 20) before incubation with 100 μl of test serum sample at a 1/1,000 dilution (in HBS-Tween) for 1 h at RT. Wells were then washed four times, with HBS replacing HBS-Tween in the final wash. Plates were then incubated for 1 h at RT with 100 μl of horseradish peroxidase-conjugated rabbit anti-human IgG (Dako) at a 1/5,000 dilution in HBS-Tween before the final wash and detection with H2O2 and o-phenylenediamine (Sigma). The reaction was stopped with 25 μl of 2 M H2SO4 per well, and absorbance was read at 492 nm. Identical positive controls (hyperimmune sera) were run in duplicate on each day of the experiment, on each plate, to allow for standardization of day-to-day and plate-to-plate variations. Twenty sera from UK residents, never exposed to malaria, were used as negative controls.
Assay of growth inhibitory activity
3D7 and FVO strain parasites were provided to the Jenner Institute by the PATH MVI GIA Reference Center. Dd2 parasites were provided by Bob Pinches (University of Oxford, UK). GB4, Camp and 7G8 parasites were provided by MR4 (reagents MRA-925, MRA-328 and MRA-152 respectively; MR4, ATCC). The assays of GIA were all performed at the Jenner Institute, Oxford University using the method of the MVI PATH GIA Reference Center7, except for the experiments reported in Figure 3 that were performed using the same method at the GIA Reference Center. Total IgG was purified using Protein G (Pierce). Briefly, each test IgG (at the concentration specified on each figure) was incubated with synchronized P. falciparum late trophozoites. Relative parasitemia levels were quantified by colorimetric determination of parasite lactate dehydrogenase after one complete life-cycle (40–42 h for 3D7; 48 h for all other parasite strains). Results were calculated relative to growth in the presence of 10 mg ml−1 IgG from a rabbit immunized with non-malaria control vaccines.
Experiments depicted in Figure 2b were performed using IgG purified from pooled serum of two PfRH5FL-vaccinated rabbits and single rabbits vaccinated with each PfAMA1 vaccine. The selection of these rabbits was performed in a manner designed to underestimate the differences between PfRH5FL and PfAMA1 vaccines: PfRH5FL-vaccinated rabbits chosen exhibited GIA activity at or below the median of the PfRH5FL-vaccinated rabbits illustrated in Figure 2a, whereas the PfAMA1-bivalent-vaccinated rabbit exhibited GIA activity exceeding the median of the PfAMA1-vaccinated rabbits illustrated in Figure 2a. The GIA assays performed at NIH (Fig. 3) used IgG from the same sera as the experiments in Figure 2b, with the exception that IgG from the two PfRH5FL-vaccinated rabbits was not pooled.
Data analysis and statistics
Estimation of the concentration of total IgG necessary to induce 50% GIA (EC50) was performed using nonlinear least squares regression in Prism 5 software (GraphPad Software). For each vaccine–parasite strain combination, a four-parameter sigmoidal dose-response curve was fitted to the relationship between log10[total IgG] and percentage GIA, constraining the bottom of the curve to 0% GIA, and the top of the curve to 100% GIA. For each parasite strain, an extra sum-of-squares F-test was used to test a null hypothesis of equal EC50 for the tested IgG samples.
Additional information
How to cite this article: Douglas, A. D. et al. The blood-stage malaria antigen PfRH5 is susceptible to vaccine-inducible cross-strain neutralizing antibody. Nat. Commun. 2:601 doi: 10.1038/ncomms1615 (2012).
Change history
19 September 2013
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
Cohen, J., Nussenzweig, V., Nussenzweig, R., Vekemans, J. & Leach, A. From the circumsporozoite protein to the RTS, S/AS candidate vaccine. Hum. Vaccin. 6, 90–96 (2010).
Goodman, A. L. & Draper, S. J. Blood-stage malaria vaccines—recent progress and future challenges. Ann. Trop. Med. Parasitol. 104, 189–211 (2010).
Holder, A. A. The carboxy-terminus of merozoite surface protein 1: structure, specific antibodies and immunity to malaria. Parasitology 136, 1445–1456 (2009).
Remarque, E. J., Faber, B. W., Kocken, C. H. & Thomas, A. W. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 24, 74–84 (2008).
Lyon, J. A. et al. Protection induced by Plasmodium falciparum MSP1(42) is strain-specific, antigen and adjuvant dependent, and correlates with antibody responses. PLoS One 3, e2830 (2008).
Dutta, S. et al. High antibody titer against apical membrane antigen-1 is required to protect against malaria in the Aotus model. PLoS One 4, e8138 (2009).
Miura, K. et al. Anti-apical-membrane-antigen-1 antibody is more effective than anti-42-kilodalton-merozoite-surface-protein-1 antibody in inhibiting plasmodium falciparum growth, as determined by the in vitro growth inhibition assay. Clin. Vaccine Immunol. 16, 963–968 (2009).
Thera, M. A. et al. A field trial to assess a blood-stage malaria vaccine. N Engl. J. Med. 365, 1004–1013 (2011).
Draper, S. J. et al. Effective induction of high-titer antibodies by viral vector vaccines. Nature Med. 14, 819–821 (2008).
Forbes, E. K. et al. Combining liver- and blood-stage malaria viral-vectored vaccines: investigating mechanisms of CD8+ T cell interference. J. Immunol. 187, 3738–3750 (2011).
Douglas, A. D. et al. Tailoring subunit vaccine immunogenicity: maximizing antibody and T cell responses by using combinations of adenovirus, poxvirus and protein-adjuvant vaccines against Plasmodium falciparum MSP1. Vaccine 28, 7167–7178 (2010).
Biswas, S. et al. Transgene optimization, immunogenicity and in vitro efficacy of viral vectored vaccines expressing two alleles of plasmodium falciparum AMA1. PLoS One 6, e20977 (2011).
Goodman, A. L. et al. New candidate vaccines against blood-stage Plasmodium falciparum malaria: prime-boost immunization regimens incorporating human and simian adenoviral vectors and poxviral vectors expressing an optimized antigen based on merozoite surface protein 1. Infect. Immun. 78, 4601–4612 (2010).
Draper, S. J. et al. Enhancing blood-stage malaria subunit vaccine immunogenicity in rhesus macaques by combining adenovirus, poxvirus, and protein-in-adjuvant vaccines. J. Immunol. 185, 7583–7595 (2010).
Sheehy, S. H. et al. Phase Ia clinical evaluation of the plasmodium falciparum blood-stage antigen MSP1 in ChAd63 and MVA vaccine vectors. Mol. Ther.; Epub 23 Aug 2011, doi:2010.1038 (2011).
Collins, W. E. et al. Efficacy of vaccines containing rhoptry-associated proteins RAP1 and RAP2 of Plasmodium falciparum in Saimiri boliviensis monkeys 62, 466–479 (2000).
Kushwaha, A., Rao, P. P., Suresh, R. P. & Chauhan, V. S. Immunogenicity of recombinant fragments of Plasmodium falciparum acidic basic repeat antigen produced in Escherichia coli. Parasite Immunol. 23, 435–444 (2001).
Cowman, A. F. & Crabb, B. S. Invasion of red blood cells by malaria parasites. Cell 124, 755–766 (2006).
Gilson, P. R. et al. Identification and stoichiometry of GPI-anchored membrane proteins of the human malaria parasite plasmodium falciparum. Mol. Cell. Proteomics 5, 1286–1299 (2006).
Jiang, L. et al. Evidence for erythrocyte-binding antigen 175 as a component of a ligand-blocking blood-stage malaria vaccine. Proc. Natl Acad. Sci. USA 108, 7553–7558 (2011).
Lopaticki, S. et al. Reticulocyte and erythrocyte binding-like proteins function cooperatively in invasion of human erythrocytes by malaria parasites. Infect. Immun. 79, 1107–1117 (2011).
Riglar, D. T. et al. Super-resolution dissection of coordinated events during malaria parasite invasion of the human erythrocyte. Cell Host Microbe 9, 9–20 (2011).
Florens, L. et al. A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526 (2002).
Baum, J. et al. Reticulocyte-binding protein homologue 5—an essential adhesin involved in invasion of human erythrocytes by Plasmodium falciparum. Int. J. Parasitol. 39, 371–380 (2009).
Tham, W. H. et al. Antibodies to reticulocyte binding protein-like homologue 4 inhibit invasion of Plasmodium falciparum into human erythrocytes. Infect. Immun. 77, 2427–2435 (2009).
Duraisingh, M. T. et al. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 22, 1047–1057 (2003).
Gao, X. et al. Antibodies targeting the PfRH1 binding domain inhibit invasion of Plasmodium falciparum merozoites. PLoS Pathog. 4, e1000104 (2008).
Pandey, K. C. et al. Bacterially expressed and refolded receptor binding domain of Plasmodium falciparum EBA-175 elicits invasion inhibitory antibodies. Mol. Biochem. Parasitol. 123, 23–33 (2002).
Rodriguez, M., Lustigman, S., Montero, E., Oksov, Y. & Lobo, C. A. PfRH5: a novel reticulocyte-binding family homolog of plasmodium falciparum that binds to the erythrocyte, and an investigation of its receptor. PLoS One 3, e3300 (2008).
Sauerwein, R. W., Roestenberg, M. & Moorthy, V. S. Experimental human challenge infections can accelerate clinical malaria vaccine development. Nat. Rev. Immunol. 11, 57–64 (2011).
Spring, M. D. et al. Phase 1/2a study of the malaria vaccine candidate apical membrane antigen-1 (AMA-1) administered in adjuvant system AS01B or AS02A. PLoS One 4, e5254 (2009).
Duncan, C. J. et al. Impact on malaria parasite multiplication rates in infected volunteers of the protein-in-adjuvant vaccine AMA1-C1/Alhydrogel+CPG 7909. PLoS One 6, e22271 (2011).
Hayton, K. et al. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe 4, 40–51 (2008).
Lobo, C. A., Rodriguez, M., Struchiner, C. J., Zalis, M. G. & Lustigman, S. Associations between defined polymorphic variants in the PfRH ligand family and the invasion pathways used by P. falciparum field isolates from Brazil. Mol. Biochem. Parasitol. 149, 246–251 (2006).
Rayner, J. C., Vargas-Serrato, E., Huber, C. S., Galinski, M. R. & Barnwell, J. W. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J. Exp. Med. 194, 1571–1581 (2001).
Crosnier, C. et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature doi: 10.1038/nature10606. [Epub ahead of print] (2011).
Gomez-Escobar, N. et al. Erythrocyte invasion and merozoite ligand gene expression in severe and mild Plasmodium falciparum malaria. J. Infect. Dis. 201, 444–452 (2010).
Mackinnon, M. J. et al. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog. 5, e1000644 (2009).
Takala, S. L. et al. Short report: rare Plasmodium falciparum merozoite surface protein 1 19-kda (msp-1(19)) haplotypes identified in Mali using high-throughput genotyping methods. Am. J. Trop. Med. Hyg. 76, 855–859 (2007).
Volkman, S. K. et al. A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 39, 113–119 (2007).
Jeffares, D. C. et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat. Genet. 39, 120–125 (2007).
Freeman, R. R., Trejdosiewicz, A. J. & Cross, G. A. Protective monoclonal antibodies recognising stage-specific merozoite antigens of a rodent malaria parasite. Nature 284, 366–368 (1980).
Crompton, P. D. et al. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc. Natl Acad. Sci. USA 107, 6958–6963 (2010).
Fowkes, F. J., Richards, J. S., Simpson, J. A. & Beeson, J. G. The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: a systematic review and meta-analysis. PLoS Med. 7, e1000218 (2010).
Gardner, M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002).
Oeuvray, C. et al. Merozoite surface protein-3: a malaria protein inducing antibodies that promote Plasmodium falciparum killing by cooperation with blood monocytes. Blood 84, 1594–1602 (1994).
Coppel, R. L. Vaccinating with the genome: a Sisyphean task? Trends Parasitol. 25, 205–212 (2009).
Triglia, T. et al. Identification of proteins from Plasmodium falciparum that are homologous to reticulocyte binding proteins in Plasmodium vivax. Infect. Immun. 69, 1084–1092 (2001).
Alcock, R. et al. Long-term thermostabilization of live poxviral and adenoviral vaccine vectors at supraphysiological temperatures in carbohydrate glass. Sci. Transl. Med. 2, 19ra12 (2010).
Reyes-Sandoval, A. et al. Prime-boost immunization with adenoviral and modified vaccinia virus ankara vectors enhances the durability and polyfunctionality of protective malaria CD8+ T-cell responses. Infect. Immun. 78, 145–153 (2010).
Sridhar, S. et al. Single-dose protection against Plasmodium berghei by a simian adenovirus vector using a human cytomegalovirus promoter containing intron A. J. Virol. 82, 3822–3833 (2008).
Stanton, R. J., McSharry, B. P., Armstrong, M., Tomasec, P. & Wilkinson, G. W. G. Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function. Biotechniques 45, 659–662 (2008).
Draper, S. J. et al. Recombinant viral vaccines expressing merozoite surface protein-1 induce antibody- and T cell-mediated multistage protection against malaria. Cell Host Microbe 5, 95–105 (2009).
Kaneko, O., Mu, J., Tsuboi, T., Su, X. & Torii, M. Gene structure and expression of a Plasmodium falciparum 220-kDa protein homologous to the Plasmodium vivax reticulocyte binding proteins. Mol. Biochem. Parasitol. 121, 275–278 (2002).
Bushell, K. M., Sollner, C., Schuster-Boeckler, B., Bateman, A. & Wright, G. J. Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res. 18, 622–630 (2008).
de Cassan, S. C. et al. The requirement for potent adjuvants to enhance the immunogenicity and protective efficacy of protein vaccines can be overcome by prior immunization with a recombinant adenovirus. J. Immunol. 187, 2602–2616 (2011).
Marsh, K. & Kinyanjui, S. Immune effector mechanisms in malaria. Parasite Immunol. 28, 51–60 (2006).
Osier, F. H. et al. Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infect. Immun. 76, 2240–2248 (2008).
Miura, K. et al. Development and characterization of a standardized ELISA including a reference serum on each plate to detect antibodies induced by experimental malaria vaccines. Vaccine 26, 193–200 (2008).
Acknowledgements
We acknowledge the assistance of the Jenner Institute Vector Core Facility and Adjuvant Bank (University of Oxford, UK). We thank Bijender Singh and Chetan Chitnis (ICGEB, New Delhi, India) for provision of PfEBA175_F2 and PfAMA1 protein; Samuel Moretz, Hong Zhou and Ababacar Diouf (PATH MVI GIA Reference Center, NIAID, NIH, USA) for technical support in performing the GIA assays at the Reference Center; Robert Pinches (WIMM, University of Oxford, UK) for the provision of Dd2 strain parasites; MR4 for providing us with Camp, 7G8 and GB4 malaria parasites originally contributed by L Miller & D Baruck, D Walliker, and K Hayton & T Wellems, respectively. We also acknowledge the assistance of Matthew Cottingham, Matthew Dicks, Prateek Choudhary and Julie Furze (Jenner Institute, Oxford, UK). A.D.D., J.J.I., G.K., F.H.O., A.V.T., K.M., C.C., G.J.W. and A.V.S.H. received Wellcome Trust support (grant numbers 089455/2/09/z; 089833/Z/09/Z; 077108; 084113/z/07/z). A.L.G. and S.J.D. received M.R.C. support (grant numbers G0600424 and G1000527). S.B. received funding from MalParTraining (an EU FP6-funded Marie Curie Action under contract number MEST-CT-2005-020492); D.H.W. received a grant from the Foundation for the NIH through the Grand Challenges in Global Health Initiative of the Gates Foundation; C.A.L. and K. Miura are supported by the PATH Malaria Vaccine Initiative (MVI) and the Intramural Program of NIH NIAID; A.V.S.H. and S.J.D. are Jenner Investigators.
Author information
Authors and Affiliations
Contributions
A.D.D., A.R.W., J.J.I., G.K., S.B., A.L.G., D.H.W., C.C., G.J.W., C.A.L., F.H.O., K. Miura, K. Marsh, A.V.S.H. and S.J.D. conceived, planned and designed experiments, including design of viral vectors and clinical trials that provided human sera. A.D.D., A.R.W., J.J.I., G.K., S.B., A.L.G., D.H.W., C.C., K Miura, G.J.W., F.H.O., A.V.T. and S.J.D. conducted experiments. A.D.D., A.R.W., J.J.I., F.H.O., K Miura, C.A.L. and S.J.D. analysed data. A.D.D., A.R.W., S.J.D.wrote the paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.D.D., A.R.W., J.J.I., A.L.G., C.C., G.J.W., A.V.S.H. and S.J.D. are named on patent applications relating to PfRH5 and/or other malaria vaccines (Treatment and Prevention of Malaria, GB1103293.5; Immunogenic Compositions, WO/2008/122817; Adenoviral Vectors Encoding a Pathogen or Tumour Antigen, WO/2008/122811; GB1016969.6).
Rights and permissions
About this article
Cite this article
Douglas, A., Williams, A., Illingworth, J. et al. The blood-stage malaria antigen PfRH5 is susceptible to vaccine-inducible cross-strain neutralizing antibody. Nat Commun 2, 601 (2011). https://doi.org/10.1038/ncomms1615
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms1615
This article is cited by
-
Malaria vaccines: the 60-year journey of hope and final success—lessons learned and future prospects
Tropical Medicine and Health (2023)
-
Recent increase in low complexity polygenomic infections and sialic acid-independent invasion pathways in Plasmodium falciparum from Western Gambia
Parasites & Vectors (2023)
-
Assessment of precision in growth inhibition assay (GIA) using human anti-PfRH5 antibodies
Malaria Journal (2023)
-
Heterotypic interactions drive antibody synergy against a malaria vaccine candidate
Nature Communications (2022)
-
Structure-guided insights into potential function of novel genetic variants in the malaria vaccine candidate PfRh5
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.