Abstract
Brown adipose tissue (BAT) dissipates nutritional energy as heat via the uncoupling protein-1 (UCP1) and BAT activity correlates with leanness in human adults. Here we profile G protein-coupled receptors (GPCRs) in brown adipocytes to identify druggable regulators of BAT. Twenty-one per cent of the GPCRs link to the Gq family, and inhibition of Gq signalling enhances differentiation of human and murine brown adipocytes. In contrast, activation of Gq signalling abrogates brown adipogenesis. We further identify the endothelin/Ednra pathway as an autocrine activator of Gq signalling in brown adipocytes. Expression of a constitutively active Gq protein in mice reduces UCP1 expression in BAT, whole-body energy expenditure and the number of brown-like/beige cells in white adipose tissue (WAT). Furthermore, expression of Gq in human WAT inversely correlates with UCP1 expression. Thus, our data indicate that Gq signalling regulates brown/beige adipocytes and inhibition of Gq signalling may be a novel therapeutic approach to combat obesity.
Similar content being viewed by others
Introduction
Brown adipose tissue (BAT) is important for basal and inducible energy expenditure in mammals1,2,3,4,5. Importantly, recent findings indicate that adult humans possess metabolically active BAT6 and that obese subjects have reduced activity of BAT7. BAT uniquely expresses the uncoupling protein-1 (UCP1), a mitochondrial protein which uncouples ATP production from oxidative phosphorylation leading to energy expenditure in a thermogenic manner. In contrast, white adipose tissue (WAT) is mainly responsible for energy storage and is an important endocrine tissue that releases adipokines, which in turn regulate energy intake and expenditure8. Brown-fat like cells have recently been described in WAT depots9 and their number and activity can be strongly induced by cold exposure, a process described as ‘browning’2,10.
G protein-coupled receptors (GPCRs) are a large family of seven transmembrane proteins11,12 that regulate important biological processes in diverse tissues including adipose tissue13,14. Approximately 25% of currently marketed drugs target GPCRs, illustrating their importance in disease and therapeutics. GPCRs are coupled to heterotrimeric G proteins, which are composed of Gα, β and γ subunits. Activation of GPCRs leads to the disassociation of Gα from the Gβγ dimer, allowing the binding and regulation of signalling effectors. The downstream signalling of GPCRs is in part determined by their G protein coupling15. There are four main classes of Gα proteins: Gs, Gi, Gq and G12/13. Activation of Gs and Gi leads to the stimulation or inhibition of cAMP signalling, respectively, while Gq activates phospholipase C (PLC). G12/13 activates the small GTPase Rho, a pathway also known to be modulated by the Gq family of proteins16,17.
Until now, the analysis of BAT GPCRs has focused mainly on a few Gs-coupled receptors (for example, β-adrenergic4 and adenosine18 receptors) that activate cAMP signalling and UCP1-dependent thermogenesis. The function of other Gα GPCR families in adipocytes is not as clear. Therefore, in the present study, we investigate the expression pattern of non-odorant GPCRs in brown adipocytes (BAs). Our analysis reveals that Gq-coupled receptors are highly expressed in brown adipocytes. Moreover, using pharmacological and genetic approaches, we found that Gq, via modulation of RhoA signalling, regulates adipogenesis of BAs in vitro and in vivo.
Results
GPCR expression of brown adipocytes
To identify the expression pattern of non-odorant/tastant GPCRs in BAs, we profiled the transcript levels of 347 GPCRs. We detected 182 GPCRs in preadipocytes and 230 in mature BAs (Supplementary Tables 1,2). 18 GPCRs and 66 GPCRs were uniquely expressed in preadipocytes and mature BAs, respectively (Fig. 1a). We used the IUPHAR database (http://www.guidetopharmacology.org/)19 to define GPCRs. Class A and adhesion GPCRs represented the two largest classes in both pre- and mature BAs (Table 1). During differentiation, class A receptors increased from 139 to 188 receptors, whereas 13 out of the 20 adhesion GPCRs expressed in preadipocytes decreased upon differentiation with five of them decreasing below the detection limit (Table 1). With respect to coupling to heterotrimeric G proteins, we found that 14.0 and 31.5% of the GPCRs detected in BAs interact with Gs and Gi, respectively (Fig. 1b). Unexpectedly, 21.5% of the GPCRs expressed in BAs are predicted to link with Gq/11 (Fig. 1b). Preadipocytes exhibited a similar pattern of GPCR linkage (Fig. 1c). During differentiation, the expression levels of 91 GPCRs changed more than twofold, with 26 and 65 showing a decrease or increase in expression, respectively (Supplementary Fig. 1a,b). During brown adipogenesis, Gq- and Gi-coupled GPCRs were the largest group of receptors with known G protein linkages to be down- and upregulated, respectively (Supplementary Fig. 1a,b).
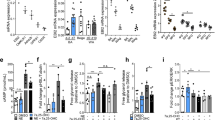
(a) Venn diagram showing unique and overlapping GPCRs expressed in murine brown preadipocytes and mature murine BA. (b) Pie chart of GPCR-linkage classification in BA. (c) Pie chart of GPCR-linkage classification in brown preadipocytes. (d) Oil Red O stain of BA chronically treated with and without FR and after knockdown of Gq with shRNA (shGq); Ctrl, control. Scale bar, 1 cm. (e–h) Representative immunoblots (e,g) and quantification (f,h) of adipogenic markers (aP2; PPARγ) and thermogenic marker (UCP1) of BA after indicated treatment. ANOVA, *P<0.05. (i,j) Representative traces of label-free DMR (i) and concentration-effect curve (j) in cells stably expressing Gq DREADD (Dq) treated with indicated concentrations of CNO. (k,l) Representative immunoblots (k) and quantification (l) of aP2, PPARγ and UCP1 in BA transduced with control virus (LVctrl) or virus containing Dq and differentiated in presence or absence of CNO. ANOVA, *P<0.05. (m,n) Representative immunoblots (m) and quantification (n) of aP2, PPARγ and UCP1 in BA transduced with control virus (LVGFP) or virus expressing constitutively active Gq (LVGqQL). (a–c), n=3 independent arrays. (d–h) n=4. (i,j) n=3. (k-l) n=4. (m,n) n=5. ANOVA, *P<0.05. All data are shown as mean±s.e.m.
To validate the expression pattern of GPCRs, we focused on six Gq-coupled GPCRs that are highly expressed in pre- and/or mature BAs as identified by the GPCR array (Supplementary Tables 1,2): Endothelin (ET) receptors type A and B (Ednra and Ednrb), Adrenoceptor Alpha 1A (Adra1a), Angiotensin II receptor, type 1a and 1b (Agtr1a and Agtr1b) and Cholinergic Receptor, Muscarinic 3 (Chrm3). Both quantitative PCR (qPCR) and TaqMan GPCR array data demonstrated high expression of Ednra mRNA in preadipocytes and an increase of Ednra during differentiation (Supplementary Fig. 1c). Ednrb increased 280-fold (qPCR) and 332-fold (GPCR array) during differentiation; Adra1a increased 83-fold (qPCR) and 322-fold (GPCR array); Agtr1a increased 3.9-fold (qPCR) and 2.9-fold (GPCR array) (Supplementary Fig. 1c).
Gq and G11 are both expressed in preadipocytes, BAs and BAT and their levels of expression did not change significantly during differentiation (Supplementary Fig. 1d). Thus, GPCR/Gq/11 signalling might contribute to the differentiation and function of BAs; however, virtually no information exists regarding this signalling pathway in BAT.
Regulation of adipogenesis by Gq
We inhibited Gq/11 signal transduction using pharmacological and genetic approaches. Treatment with FR900359 (FR; 1 μM), a natural inhibitor of Gq/11 (ref. 20), enhanced differentiation of murine BAs, as demonstrated by increased Oil Red O staining (Fig. 1d) and elevated protein levels of the adipogenic markers aP2 and PPARγ as well as of the thermogenic marker UCP1 (Fig. 1e,f). To complement the pharmacological approach, we used lentiviral small hairpin RNA (shRNA) directed only against Gq (shGq). Knockdown of Gq by ∼50% (Supplementary Fig. 1e) significantly increased adipogenesis, with increased lipid staining (Fig. 1d) and expression of adipogenic markers and UCP1 (Fig. 1g,h). Inactivation of Gq signalling thus promotes differentiation of murine BAs.
To determine the impact of increased Gq signalling on brown adipogenesis, we first selectively enhanced Gq signalling using a Gq-coupled designer GPCR (DREADD)21 that represents a modified M3 muscarinic receptor (Dq)22. Dq couples exclusively to Gq and is activated by otherwise pharmacologically inactive clozapine-N-oxide (CNO)22. Brown preadipocytes were transduced with lentiviral vectors expressing Dq. To analyse Gq-induced responses in living cells in a label-free manner, we used biosensor technology based on dynamic mass redistribution (DMR). Treatment of Dq-expressing cells with CNO (1 μM) induced a concentration-dependent DMR response with half-maximal activation at 269 nM (Fig. 1i,j). In contrast, no DMR response was observed in buffer- or CNO-treated wild-type cells (Supplementary Fig. 1f). The Dq-induced DMR response was markedly different from the optical traces obtained by stimulation of Gs with isoprenaline (Supplementary Fig. 1g,h), implying that DMR can be used to monitor different G protein signalling pathways in BAs. Treatment of Dq cells with FR abolished the CNO-induced DMR response, providing further evidence for FR as a Gq inhibitor (Fig. 1j and Supplementary Fig. 1i).
Dq stimulation during BA differentiation reduced Oil Red O staining (Supplementary Fig. 1j), suppressed expression of adipogenic markers aP2 and PPARγ and of UCP1 (Fig. 1k,l). Treatment of non-transduced control cells or cells transduced with a control virus (LVctrl) with CNO did not affect differentiation (Fig. 1k,l and Supplementary Fig. 1j). Expression of Dq without CNO stimulation did not alter BA differentiation (Fig. 1k,l and Supplementary Fig. 1j).
Secondly, we used lentiviral vectors to express a constitutively active mutant of Gq (LVGqQL) as a means to enhance Gq signalling. We validated the activity of GqQL in brown preadipocytes by measuring IP1 in non-transduced cells and cells that express either GFP or GqQL. IP1 is a downstream metabolite of PLC and IP3, a pathway known to be activated by Gq GPCRs. We found that IP1 levels were highly increased in preadipocytes that express GqQL compared with GFP-expressing or control cells (Supplementary Fig. 1k). GqQL overexpression during adipocyte differentiation reduced Oil Red O staining and inhibited the expression of adipogenic and thermogenic markers (Fig. 1m,n and Supplementary Fig. 1l) in a similar manner as did the DREADD approach.
Endothelin/Ednra regulation of brown adipocytes
To identify an endogenous receptor that activates Gq signalling in BAs, we focused on Ednra, one of the most highly expressed Gq-coupled GPCRs in preadipocytes and BAs (Supplementary Tables 1,2). Both preadipocytes and BAs express constituents of the ET system: Endothelin-1 (ET-1), ET receptors type A and B (Ednra and Ednrb) and ET converting enzyme-1 (ECE-1) (Supplementary Fig. 2a). Although ECE-1 was expressed at low levels in BA, it is not the only enzyme that can catalyse the production of ET-1 (ref. 23). Importantly, ET-1 was released by both cell types (Fig. 2a) and its secretion was inhibited by noradrenaline (NE) (Fig. 2a).
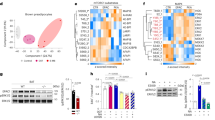
(a) Release of ET-1 by preadipocytes, non-stimulated and NE-stimulated BA. ANOVA, *P<0.05. (b) Oil Red O stain of BA chronically treated with ET-1, Ednra inhibitor BQ-123, or both. Scale bar, 1 cm. (c,d) Representative immunoblots (c) and quantification (d) of adipogenic markers and UCP1 after treatment of BA with and without ET-1, BQ-123 or both. ANOVA, *P<0.05. (e) Oil Red O stain of BA chronically treated with ET-1, FR, or both. Scale bar, 1 cm. (f,g) Representative immunoblots (f) and quantification (g) of aP2, PPARγ and UCP1 of BA chronically treated with and without ET-1, FR or both. ANOVA, *P<0.05. (h) Phalloidin staining of F-Actin stress fibres in non-transduced cells and cells transduced with control virus (LVGFP) or virus expressing constitutively active Gq (LVGqQL) in serum-starved and stimulated state. Scale bar, 50 μm. (i) RhoA activation assay in preadipocytes expressing GFP (LVGFP) or GqQL (LVGqQL). t-test, *P<0.05. (j) Oil Red O stain of non-transduced BA and BA expressing GFP (LVGFP) or GqQL (LVGqQL) differentiated in presence or absence of ROCK inhibitor Y-27632. Scale bar, 1 cm. (k,l) Representative immunoblots (k) and quantification (l) of aP2, PPARγ and UCP1 in BA non-transduced and transduced with lentivirus expressing GFP (LVGFP) or GqQL (LVGqQL) and differentiated in presence or absence of Y-27632. (a–d) n=4. (e–i) n=3. (j–l) n=5. ANOVA, *P<0.05. All data are shown as mean±s.e.m.
Differentiation of BAs was inhibited by chronic treatment with ET-1 (0.3 nM) but was restored by the selective Ednra antagonist BQ-123 (100 nM) (Fig. 2b–d) and by treatment with FR (1 μM) (Fig. 2e–g). Notably, BQ-123 increased the differentiation of the untreated cells. In contrast, ET-1 effects on differentiation were not restored with the Ednrb antagonist BQ-788 (100 nM) (Supplementary Fig. 2b–d). Moreover, acute treatment of mature BAs with ET-1 reduces lipoprotein lipase gene expression24. Lipoprotein lipase is important for fatty acid uptake and fuels cold-/NE-induced BAT thermogenesis25. Together these data implicate ET-1 as an autocrine/paracrine inhibitor of BA function that signals via Ednra/Gq and is regulated by NE signalling.
Gq regulation of BAs via Rho/Rho-kinase
Gq can activate multiple intracellular signalling pathways including IP3/DAG, Ca2+, ERK and also by GTP-binding proteins of the Ras and Rho families16,17. Since Rho/Rho-kinase (ROCK) controls brown adipogenesis26,27, we evaluated this pathway. Cells expressing GqQL had a marked increase in phalloidin staining of actin filaments (Fig. 2h) and Rho activity (Rho-GTP pull-down assay) (Fig. 2i). In addition, pharmacological inhibition of ROCK with Y-27632 (10 μM) enhanced differentiation and increased expression of adipogenic and thermogenic markers in GqQL-expressing BAs (Fig. 2j–l). ROCK inhibition also restored adipogenesis of ET-1 treated cells (Supplementary Fig. 2e–g). These results suggest that Gq inhibits BA differentiation via the RhoA/ROCK pathway.
In vivo regulation of BAT by Gq signalling
To study the effects of elevated Gq signalling in vivo, we expressed constitutively active Gq in BAT (Fig. 3a). Lentiviral particles encoding either GqQL (LVGqQL) or GFP (LVGFP) under the control of the CMV promoter were directly injected into BAT of mice that were subsequently cold-exposed for 2 weeks (Fig. 3b). Histological analysis revealed increased size of the lipid droplets with more unilocular, white-like adipocytes in LVGqQL-injected BAT and decreased UCP1 staining (Fig. 3c). Expression of GqQL decreased UCP1 protein level in BAT by ∼20% (Fig. 3d) and decreased expression of the mitochondrial marker genes Nd5 and Ndufa (Fig. 3a).
(a) Relative mRNA expression of Gq, Ndufa and Nd5 in BAT of LVGFP and LVGqQL mice. t-test, *P<0.05. (b) Representative images of interscapular BAT of mice injected with LVGFP or LVGqQL in bright field (left) and fluorescent microscopy (right). Scale bar, 5 mm. (c) Representative hematoxylin/eosin (HE) and UCP1 staining of BAT in LVGqQL and LVGFP mice after cold stimulation. Scale bar, 100 μm. (d) Representative immunoblots (top) and quantification (bottom) of UCP1 expression in BAT of LVGFP and LVGqQL mice. t-test, *P<0.05. (e) Relative mRNA expression of Gq and UCP1 in BAT of UCP1-GFP and UCP1-GqQL transgenic mice. t-test, *P<0.05. (f) Representative HE and UCP1 staining of BAT in UCP1-GFP and UCP1-GqQL mice after cold stimulation. Scale bar, 100 μm. (g) Representative immunoblots (top) and quantification (bottom) of UCP1 expression in BAT of UCP1-GFP and UCP1-GqQL mice. t-test, *P<0.05. (h) Oxygen consumption of UCP1-GFP and UCP1-GqQL mice over 24 h. (i) Area under the curve (AUC) of the oxygen consumption of UCP1-GFP and UCP1-GqQL mice over 24 h t-test, *P<0.05. (a–d) 11 animals per group were analysed. (e,g) 6 animals per group were analysed., (f,h,i) 4 animals per group were analysed. All data are shown as mean±s.e.m.
In addition, we generated transgenic mice that express either GqQL or GFP (control) under the control of the UCP1 promoter (UCP1-GqQL and UCP1-GFP, respectively) to achieve BA-specific expression (Fig. 3e), and exposed them to cold. Similar to what occurred after injection of LVGqQL, immunohistochemical staining of BAT from UCP1-GqQL mice revealed enlarged, unilocular lipid droplets and reduced UCP1 staining (Fig. 3f). UCP1 expression of BAT of UCP1-GqQL mice was significantly decreased compared with the control UCP1-GFP group (Fig. 3e,g). Indirect calorimetry revealed significantly reduced O2 consumption in cold-exposed UCP1-GqQL mice (Fig. 3h,i). Together, these results indicate that Gq activation induces a phenotypic change of BAT towards WAT (a process also known as ‘whitening’28) with diminished thermogenic capacity of BAT.
Apart from classical interscapular BAT, inducible brown (also known as beige/brite) adipocytes have been identified in murine and human WAT2,10. Exposure of transgenic UCP1-GqQL mice to cold, induced a 1.9-fold and 1.5-fold increase in UCP1 promoter-driven Gq expression in inguinal (iWAT) and gonadal/visceral (gWAT) WAT, respectively (Supplementary Fig. 3a,b). Histological analysis of both iWAT and gWAT revealed ‘browning’ in the control mice that express GFP under the control of the UCP1 promoter (Fig. 4a and Supplementary Fig. 3c). In contrast, iWAT and gWAT of GqQL mice exhibited larger lipid droplets with decreased expression of BA markers (UCP1, PGC1-α and DIO2) (Fig. 4a,b and Supplementary Fig. 3c,d).
(a) Representative HE and UCP1 staining of inguinal WAT in UCP1-GFP and UCP1-GqQL mice after cold stimulation. Scale bar, 100 μm. (b) Relative mRNA expression of thermogenic markers in inguinal WAT of UCP1-GqQL relative to UCP1-GFP mice. t-test, *P<0.05. (c) UCP1 expression in NE-stimulated murine brown/beige-like adipocytes transduced with control virus (LVctrl) or lentivirus carrying Gq DREADD (Dq) in presence and absence of CNO. ANOVA, *P<0.05. (d) Oil Red O stain of human brown/beige adipocytes (hMADS) differentiated in the presence or absence of FR. Scale bar, 100 μm. (e) Relative mRNA expression of adipogenic markers (aP2; PPARγ) and UCP1 of hMADS differentiated in the presence or absence of FR normalized to GAPDH (Glyceraldehyde 3-phosphate dehydrogenase). ANOVA, *P<0.05. (f) Correlation of Gq and UCP1 expression in human WAT. (g) Scheme of ET-1/Gq signalling in BA differentiation and browning. (a,b) 6 animals per group were analysed. (c–e) n=3. (f) 266 human abdominal WAT samples were analysed. All data are shown as mean±s.e.m.
Further evidence for a role of Gq in inducible brown/beige cells was obtained by induction of browning in isolated primary murine adipocytes (Fig. 4c). Using the DREADD system (Fig. 1i–l), we activated Gq signalling with CNO (10 μM), which resulted in a significant reduction in UCP1 expression (Fig. 4c).
To explore the effect of enhanced Gq signalling on white adipogenesis, we used murine white adipocytes isolated from iWAT (WAi). We overexpressed Dq and activated it with CNO throughout differentiation. Gq activation suppressed WAi differentiation (Supplementary Fig. 3e–g).
Role of Gq signalling in human adipocytes
Human BAT also highly expresses Gq protein. We found Gq-coupled receptors that were highly expressed in murine BAs are also highly expressed in the human brown/beige adipocyte cell line (hMADS) (Ednra, Ednrb, Adra1a, Agtr1 and Chrm3) (Supplementary Fig. 3h,i). Treatment of the hMADS cells with FR (1 μM) enhanced differentiation and expression of adipogenic markers aP2 and PPARγ and thermogenic marker UCP1 (Fig. 4d,e). Moreover, we found a relationship between expression of Gq and the thermogenic markers UCP1 and CIDEA in WAT samples from 266 human subjects (Fig. 4f and Supplementary Fig. 3j). UCP1 and CIDEA expression were both significantly inversely correlated with Gq; results that are consistent with the idea that Gq expression negatively affects the thermogenic capacity in humans (Fig. 4f and Supplementary Fig. 3j).
Discussion
In the present study, we screened for non-chemosensory GPCRs in murine brown preadipocytes and mature adipocytes. GPCR profiling revealed that BAs express more than 180 GPCRs and that the expression pattern changes greatly upon differentiation. The number of GPCRs detected is in agreement with previous analysis performed on human subcutaneous adipose tissue and murine metabolic tissues that detected 163 and 198 GCPRs, respectively29,30. In BAs, we found that Gq-coupled receptors were one of the largest groups and that their expression was highly altered during differentiation, indicating a potential role in adipogenesis. Importantly, we show that Gq signalling is a crucial inhibitor of adipocyte differentiation, through activation of the Rho/ROCK signalling cascade, a major regulator of BA differentiation26 (Fig. 4g).
Treatment with the Gq inhibitor FR enhanced the adipogenic and thermogenic potential of BAs, indicating either the presence of constitutively active Gq receptors and/or an autocrine loop of Gq signalling in BAs. Analysis of highly expressed Gq-coupled GPCRs indicated that murine BAs and human brown/beige cells express ET receptors. Moreover, murine BAs also express the enzymes necessary for ET production and secretion. Stimulation with ET-inhibited adipogenesis, while the inhibition of Gq and Ednra, but not Ednrb, rescued adipogenesis of ET-treated cells. Recently, Xue et al.31 identified Endrb as a genetic marker that positively correlates with the thermogenic capacity of cloned human BAs. Inhibition of Ednra alone increased the differentiation of untreated cells, indicating endogenous activity of the ET system in BAs. Thus, our data implicate the ET-1/Ednra signalling axis as an autocrine/paracrine inhibitor of brown/beige adipogenesis via Gq signalling (Fig. 4g). In addition to its function in BAT, ET-1 plays an important role in the vasculature and excessive ET-1 contributes to atherosclerosis32 as well as to the disturbed insulin-induced vasorelaxation in obesity33.
Our data also indicate an inhibitory role of Gq in BAT function and browning in vivo. Overexpression of GqQL induced a phenotype of BAT reminiscent of ‘whitening’, while preventing the ‘browning’ of WAT in mice exposed to cold. Overall, activation of Gq signalling in brown/beige adipocytes resulted in reduced whole-body energy expenditure of cold-exposed mice. Thus, our data show that the Gq signalling pathway inhibits brown and beige adipose tissue in vivo and the studies done using cultured adipocytes show that Gq signalling inhibits adipogenesis in a more general manner. Furthermore, we found a negative correlation between Gq and UCP1 mRNA expression in human abdominal WAT. Although abdominal adipose tissue is predominantly white, it can be induced to brown and inducible brown-like cells have been identified in abdominal fat after β-3 adrenergic receptor treatment34. Further studies are required to investigate the role of alterations in the expression of BAT marker genes in abdominal fat depots in adult humans. Interestingly, Gq signalling is not only involved in regulation of adipocyte differentiation and non-shivering thermogenesis, but also in the CNS regulation of appetite35.
In conclusion, our results demonstrate a previously unappreciated role in brown/beige adipogenesis for Gq, to which ∼20% of the GPCRs expressed in these cells are predicted to couple. Our findings suggest that antagonizing Gq signalling by targeting Gq-coupled GPCRs and/or by directly inhibiting Gq represent previously unexplored ways to enhance the amount of brown/beige fat and thus increase energy expenditure.
Methods
Isolation and differentiation of BAs
BAT-derived mesenchymal stem cells were isolated from interscapular BAT of newborn wild-type mice26,36. Excised BAT was incubated for 30 min at 37 °C in digestion buffer (Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) containing 123 mM Na+, 5 mM K+, 1.3 mM Ca2+, 131 mM Cl−, 5 mM glucose, 1.5% (w/v) bovine serum albumin (BSA), 100 mM Hepes and 0.2% (w/v) collagenase type II (pH 7.4)). After digestion, tissue remnants were removed by filtration through a 100 μm nylon mesh and placed on ice for the next 30 min. The infranatant was filtered through a 30 μm nylon mesh and centrifuged at 700g for 10 min. The pellet containing mesenchymal stem cells was resuspended in DMEM supplemented with 10% fetal bovine serum (FBS), 100 IU penicillin, streptomycin (100 μg ml−1) (P/S), 4 nM insulin, 4 nM triiodothyronine, 10 mM Hepes and sodium ascorbate (25 μg ml−1). Approximately 60,000 cells per cm2 were seeded on a six-well plate and grown at 37 °C and 5% CO2. Preadipocytes were immortalized using the lentivirus containing the SV40 large T antigen and unselected cells were cultured in DMEM supplemented with FBS and P/S (GM). The cells were expanded in GM at 37 °C, 5% CO2 (ref. 37).
For differentiation, immortalized cells were seeded in GM on a six-well plate in a density of ∼180,000 cells per well. 48 h after seeding (day −2), GM was replaced by differentiation medium (DM), supplemented with FBS, P/S, 20 nM insulin and 1 nM triiodothyronine. Confluent cells (day 0) were then treated for 48 h with BAT induction medium, which was DM supplemented with 0.5 mM isobutylmethylxanthine (IBMX) and 1 μM dexamethasone. After the induction, cells were treated with DM for the next 5 days, which was replenished every second day. Where indicated, cells were treated every second day with the Gq inhibitor FR (1 μM), ROCK inhibitor Y-27632 (Tocris, 10 μM), CNO (Tocris, 1 μM), Endothelin-1 (ET-1) (Tocris, 0.3 nM), Ednra inhibitor BQ-123 or Ednrb inhibitor BQ-788 (both Tocris, 100 nM). In the experiments with ET-1; FR, Y-27632, BQ-123 or BQ-788 were added to the culture medium 30 min before the addition of ET-1.
For all the preadipocyte experiments, the cells were used on day −2. For mature BA experiments, the cells were used on day +7 after induction.
Isolation and differentiation of white and beige adipocytes
WAT-derived mesenchymal stem cells were isolated from inguinal white fat pads of 8- to 12-week-old wild-type mice. Excised WAT was incubated for 30 min at 37 °C in digestion buffer (DMEM containing 0.5% (w/v) BSA and 0.15% (w/v) collagenase type II). After digestion, cells were allowed to stand for 10 min at room temperature and centrifuged at 700g for 10 min. The pellet was resuspended in GM and filtered through a 100 μm nylon mesh. Approximately 180,000 cells per well were seeded in GM on a six-well plate at 37 °C and 5% CO2.
For differentiation, cells were induced 2 days after reaching confluence (day 0) for 48 h, using WAT induction medium (DMEM supplemented with 5% FBS, 1% P/S, 1 μM dexamethasone, 0.5 mM IBMX, 1 nM triiodothyronine, 1 mM D-biotin, 17 mM pantothenate, L-ascorbate (50 mg ml−1), 1 μM rosiglitazone and 0.172 mM insulin). After the induction, cells were treated with maintainance medium (DMEM supplemented with 5% FBS, P/S, 1 nM triiodothyronine, 1 mM D-biotin, 17 mM pantothenate, L-ascorbate (50 mg ml−1), 1 μM and 0.172 μM insulin) for the next 7 days, which was replenished every second day. For differentiation experiments, cells were treated every second day with CNO (1 μM). Browning was induced by 8 h NE (1 μM) stimulation, in the presence or absence of CNO (10 μM).
For all the preadipocyte experiments, the cells were used on day −2. For mature white adipocyte experiments, the cells were used on day +7 after induction.
hMADS differentiation
Human multipotent adipose-derived stem cells were provided by the laboratory of C. Dani (University of Nice SophiaAntipolis)38 and differentiated into brown/beige cells as follows18,39. Cells were seeded on 12-well plates in growth medium (DMEM Low Glucose (Lonza), supplemented with 1x glutamine (Lonza), 10 mM Hepes buffer (Lonza), penicillin-streptomycin 5,000 IU ml−1 to 5,000 UG ml−1 (Lonza) and 10% FBS (S.A Dutscher, Brumath, France)) containing 2.5 ng ml−1 FGF2 (Peprotech) at a density of ∼160,000 cells per well. After 48 h, medium was replaced with growth medium containing no FGF2. When the cells reached confluency, growth medium was replaced by hMADS induction medium (day 0) (growth medium supplemented with 5 μg ml−1 insulin, 10 μg ml−1 transferrin, 0.2 nM triiodothyronine, 1 μM rosiglitazone, 100 μM IBMX and 1 μM dexamethasone) for the next 72 h. Cells were then cultured in differentiation medium (induction medium without IBMX and dexamethasone) for 9 more days. Where indicated, cells were treated from day 0 to day 12 with the Gq inhibitor FR (1 μM).
For all the pre-adipocyte experiments, the cells were used on day −2. For mature BA experiments, the cells were used on day +12 after induction.
RNA isolation and real-time RT-PCR (qPCR)
RNA was isolated using Trizol method (Analytik Jena AG). The samples from human retroperitoneal BAT have been previously described40. cDNA was synthesized from 0.5 μg RNA using Transcriptor First Strand cDNA Synthesis Kit (Roche). Real-time RT-PCR (qPCR) was performed with SYBR-Green PCR master mix (Applied Biosystems) or LightCycler 480 SYBR Green I Master (Roche) using a HT7900 instrument (Applied Biosystems). Fold changes were calculated using relative quantification methods with mHPRT (murine hypoxanthine guanine phosphoribosyl transferase) serving as an internal control unless otherwise stated. Primer sequences are available in Supplementary Table 3.
GPCR expression analysis
GPCR profiling was performed using TaqMan Mouse GPCR Arrays (Applied Biosystems) for quantitative expression analysis of mouse GPCR genes41. These arrays detect 347 non-chemosensory GPCRs. cDNA was synthesized from 0.5 μg RNA using Transcriptor First Strand cDNA Synthesis Kit (Roche). Two nanograms of cDNA was used for each gene in the GPCR arrays. Quantification of GPCR cDNA expression was normalized to 18S rRNA. Data shown are an average of three independent arrays. GPCRs were considered unexpressed if at least two of the arrays had a delta Ct above 25. G protein coupling of GPCRs was determined using the IUPHAR database (http://guidetopharmacology.org/).
Lentiviral infection
Constitutively active Gq (GqQL), was provided by Silvio Gutkind, NIH42 Constitutive activity was achieved by mutating Glutamine to Leucine at the amino acid residue 209. Lentiviral vectors were obtained by cloning constitutively active Gq into the Bam HI and Sal I sites of the vector p156rrlsinPPTCMV, which carries a murine uncoupling protein-1 promoter (UCP1-GqQL) or ubiquitous CMV promoter (LVGqQL). Control vectors (p156rrlsinPPT) contain green fluorescent protein (UCP1-GFP and LVGFP).
The shRNA against Gq and the control shRNA were purchased from Sigma Aldrich and were expressed under the U6 promoter (pLKO.1-U6-sh- ctrl sequence: 5′-GCATGCAGAAGTGTAAAGCTA-3′, pLKO.1-U6-sh-Gq-287918-PGK-Puro 5′-GCTTGTGGAATGATCCTGGAA-3′).
pLenti6.3V5-HA-hM3D(Gq)-mCherry (Dq) lentiviral vectors were obtained from Bryan Roth, University of North Carolina. pLenti6.3-CMV-TO-GFP-Blasticidin was used as the control virus (LVctrl) for the Dq experiments.
Lentiviral particles were generated by transfection of HEK293T cells with vector constructs and packaging plasmids26,36. Viral particles were concentrated by ultracentrifugation26,36. For the lentiviral infection, cells were seeded on a six-well plate and transduced with lentiviruses corresponding to 50 ng (RTase quantified by ELISA) UCP1-GqQL, UCP1-GFP, LVGqQL or LVGFP; 75 ng Dq or LVctrl and 100 ng of the shRNA viruses for 12–16 h. Cells were further differentiated as described above.
Oil Red O staining
Mature BAs were fixed in PBS containing 4% paraformaldehyde (PFA). After washing with PBS, cells were incubated with Oil Red O (Sigma) solution (3 mg ml−1 in 60% isopropyl alcohol) for 3 h at room temperature, washed with PBS and visualized under a microscope.
Western blot analysis
Protein lysates from cells and tissues were isolated26,36 using lysis buffer (50 mM Tris, pH 7.5, 150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.1 mM EDTA and 0.1 mM EGTA) supplemented with complete protease inhibitor cocktail (Roche), 1 mM Na3VO4 and 10 mM NaF. Protein contents were determined by the Bradford method. Proteins were separated using SDS–polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane.. Membrane was blocked for 1 h in 5% BSA in Tris-Buffered Saline and 0.1% Tween 20 (TBST) and incubated over night at 4 °C in different primary antibodies (all primary antibodies diluted 1:1,000). Incubation in secondary antibody was performed the next day, for 1 h at room temperature in 5% milk in TBST. Proteins were visualized with an enhanced chemiluminescence (ECL) reagent and quantified by densitometric analysis with Image J software. Antibodies directed at the following were used: aP2 (Santa Cruz Biotechnology, Cat#: sc-18661), PPARγ (Santa Cruz Biotechnology, Cat#: sc-7273); UCP1 (Sigma Aldrich, Cat#: sc-6529 and Thermo Fisher Scientific, Cat#: PA1-24894); Tubulin (Dianova, Cat#: MS-719-P0) and GAPDH (Cell Signalling, Cat#: 2118). Secondary horse radish peroxidase–linked antibodies against goat (Dianova, Cat#: 705-035-147, dilution 1:5,000), mouse (Dianova, Cat#: 115-035-146, dilution 1:10,000) and rabbit (Cell Signalling, Cat#: 7074, dilution 1:5,000) were used. Complete immunoblots of western blot sections are shown in Supplementary Fig. 4.
Label-free dynamic mass redistribution assays
The DMR analysis was carried out using the Corning Epic system (Corning, NY, USA) in conjunction with a Cybi-SELMA semi-automated electronic pipetting system (Analytik Jena AG, Jena, Germany)43. Briefly, brown preadipoyctes stably expressing Dq were seeded on fibronectin-coated 384-well biosensor plates at a density of 3,000 cells per well and cultivated for 48 h in growth medium (37 °C, 5% CO2). Before the measurement, cells were washed twice with assay buffer (Hanks balanced salt solution with 20 mM HEPES). Where indicated, cells were pre-incubated with FR (1 μM) for 1 h or cholera toxin (CTX; 100 ng ml−1) for 18–20 h before measurement. DMR was monitored for at least 5,000 s. The optical DMR recordings were buffer-corrected. To quantify the DMR signals for concentration-effect curves, the maximum response within 1,000 s was calculated. pEC50 value determination and data calculation were performed using GraphPad Prism 5.04 (GraphPad Software, La Jolla, USA).
IP1 assay
Intracellular alteration of the second messenger IP1 was quantified with the HTRF-IP1 kit (Cisbio Bioassays) following the manufacturer’s instructions. Briefly, brown preadipocytes non-transduced and transduced with lentiviruses carrying GqQL (LVGqQL) or GFP (LVGFP) were cultivated for 48 h in growth medium (37 °C, 5% CO2). For the assay, cells were resuspended in stimulation buffer containing 50 mM LiCl and transferred to a 384-well microtiter plate at a density of 12,500 cells per well. After 90 min incubation time, 3 μl of IP1-d2 conjugate followed by 3 μl of europium cryptate-labelled anti-IP1antibody solved in lysis buffer were added to the cells to quantify intracellular IP levels. After a further incubation of 1 h at room temperature, time-resolved fluorescence was measured at 620 and 665 nm with the Mithras LB 940 multimode reader.
ET-1 ELISA assay
Cell culture medium was collected from the preadipocytes and mature BAs, with or without 8 h NE (1 μM) stimulation. ET-1 ELISA was performed using the Endothelin-1 ELISA kit (Enzo) according to the manufactureŕs instructions.
F-actin staining of adherent cells in culture
Glass coverslips were placed in 24-well plates and coated with fibronectin (5 μg ml−1). Brown preadipocytes were seeded at a density of 20,000 cells per well and serum-starved for 24 h. Cells were incubated for 30 min with 10% FBS to induce the formation of F-actin stress fibres. Cells were then fixed with 4% PFA, permeabilized with 0.1% Triton-100, blocked with 1% BSA/PBS and stained with phalloidin-Alexa 546. Coverslips were mounted on glass slides using PermaFluor mounting medium and visualized using a confocal microscope.
RhoA activation assay
Brown preadipocytes were seeded on a 10 cm tissue plate at a density of 180,000 cells per plate and infected with lentiviruses carrying GqQL or GFP (control) for 8 h. Cells were cultured until reaching 50–60% confluency, serum-starved for 24 h and collected for RhoA activity measurement. RhoA activation was assessed with an ELISA-based RhoA G-LISA Activation Assay Kit (Cytoskeleton). The level of RhoA activity was determined by colorimetric measurement at 490 nm. Active RhoA was normalized to total RhoA protein.
Lentiviral injections of constitutively active Gq (CMV-GqQL) into BAT
Four-week-old male mice (C57BL/6, Charles River) were anaesthetized using isoflurane. A small incision was made in the neck region and 1 μg of lentiviruses carrying either GqQL (LVGqQL) or GFP (LVGFP) under control of the ubiquitous CMV promoter were injected directly into each fat pad of BAT. After injections, mice were acclimatized to cold for 1 week at 18 °C following 1 week of cold exposure at 4 °C. During the study, the mice were maintained on a daily cycle of 12 h light (0600 to 1800 hours) and 12 h darkness (1800 to 0600 hours), and were allowed free access to standard chow and water. The study was approved by the Landesamt für Natur, Umwelt und Verbraucherschutz, NRW, Germany.
Generation of transgenic mice and energy expenditure
Transgenic mice were generated by subzonal injections44 of the lentiviruses UCP1-GqQL and UCP1-GFP into oocyte of a wild-type donor mouse with C57BL/6 background. Ten-week-old female mice were kept for one week at 18 °C followed by one week cold exposure at 4 °C. Oxygen consumption was measured with Phenomaster (TSE Systems) for 120 s every 16 min for 24 h. During the study, the mice were maintained on a daily cycle of 12 h light (0600–1800 hours) and 12 h darkness (1800–0600 hours), and were allowed free access to standard chow and water. The study was approved by the Landesamt für Natur, Umwelt und Verbraucherschutz, NRW, Germany.
Immunohistochemistry
BAT and WAT was fixed in PBS containing 4% PFA for 48 h and dehydrated using ethanol. Tissue was embedded in paraffin and cut into 5 μM sections. Sections were blocked with 2% normal goat serum-TBS (tris-buffered saline) for 30 min RT and immunohistochemical stainings were performed with a primary antibody (UCP1, 1:1,000, Sigma Aldrich) over night. Secondary antibody-conjugated with horseradish peroxidase (Santa Cruz Biotechnology) was applied for 1 h at RT and sections were visualized using DAB substrate (Vector Laboratories). Standard haematoxylin and eosin (HE) staining was performed on 5 μM tissue sections.
Analysis of human adipose tissue
Gq, UCP1 and CIDEA mRNA expression were measured in abdominal omental and adipose tissue (AT) samples obtained in parallel from 266 donors45 who underwent open abdominal surgery for Roux-en-Y bypass, sleeve gastrectomy, explorative laparotomy or elective cholecystectomy. All participants gave their written informed consent before taking part in the study. All investigations have been approved by the ethics committee of the University of Leipzig (363-10-13122010 and 017-12-230112) and were carried out in accordance with the Declaration of Helsinki. Human Gq, UCP1 and CIDEA mRNA expression was measured by qPCR using Assay-on-Demand gene expression kit (Gq: forward: 5′-GTGGAGAAGGTGTCTGCTTTTGA-3′; reverse: 5′-ATCTTGTTGCGTAGGCAGGTAGG-3′; UCP1: Hs00222453_m1; CIDEA: Hs00154455_m1; Applied Biosystems, Darmstadt, Germany), and fluorescence was detected on an ABI PRISM 7000 Sequence Detector (Applied Biosystems). Gq, UCP1 and CIDEA mRNA expression was calculated relative to the mRNA expression of HPRT1 mRNA (Hs01003267_m1; Applied Biosystems).
Statistics
For cell culture experiments, ‘n’ indicates the number of cultures grown and differentiated independently. For experiments with mice, ‘n’ indicates number of mice per each group.
Single comparisons were analysed using two-tailed student’s t-test. For RhoA activity assay, one-tailed student’s t-test was used. Multiple comparisons were analysed using analysis of variance (ANOVA) with Newman-Keuls post-hoc test. Values below 0.05 were considered significant. Analyses were performed using GraphPad Prism 5 software. All data are represented as mean±s.e.m. The sample size was chosen based on our previous in vitro and in vivo studies39.
Additional information
How to cite this article: Klepac, K. et al. The Gq signalling pathway inhibits brown and beige adipose tissue. Nat. Commun. 7:10895 doi: 10.1038/ncomms10895 (2016).
References
Pfeifer, A. & Hoffmann, L. S. Brown, beige, and white: the new color code of fat and its pharmacological implications. Annu. Rev. Pharmacol. Toxicol. 55, 207–227 (2015).
Rosen, E. D. & Spiegelman, B. M. What we talk about when we talk about fat. Cell 156, 20–44 (2014).
Harms, M. & Seale, P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263 (2013).
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004).
Kajimura, S. & Saito, M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu. Rev. Physiol. 76, 225–249 (2013).
Virtanen, K. A. et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360, 1518–1525 (2009).
van Marken Lichtenbelt, W. D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 (2009).
Galic, S., Oakhill, J. S. & Steinberg, G. R. Adipose tissue as an endocrine organ. Mol. Cell. Endocrinol. 316, 129–139 (2010).
Cinti, S. Transdifferentiation properties of adipocytes in the adipose organ. Am. J. Physiol. Endocrinol. Metab. 297, E977–E986 (2009).
Frontini, A. & Cinti, S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell. Metab. 11, 253–256 (2010).
Kobilka, B. K. Structural insights into adrenergic receptor function and pharmacology. Trends. Pharmacol. Sci. 32, 213–218 (2011).
Lefkowitz, R. J. Seven transmembrane receptors: something old, something new. Acta Physiol. (Oxf.) 190, 9–19 (2007).
Latek, D., Modzelewska, A., Trzaskowski, B., Palczewski, K. & Filipek, S. G protein-coupled receptors--recent advances. Acta. Biochim. Pol. 59, 515–529 (2012).
Wettschureck, N. & Offermanns, S. Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85, 1159–1204 (2005).
Neves, S. R., Ram, P. T. & Iyengar, R. G protein pathways. Science 296, 1636–1639 (2002).
Buhl, A. M., Johnson, N. L., Dhanasekaran, N. & Johnson, G. L. G alpha 12 and G alpha 13 stimulate Rho-dependent stress fiber formation and focal adhesion assembly. J. Biol. Chem. 270, 24631–24634 (1995).
Wang, Y., Falting, J. M., Mattsson, C. L., Holmstrom, T. E. & Nedergaard, J. In brown adipocytes, adrenergically induced beta(1)-/beta(3)-(Gs)-, alpha(2)-(Gi)- and alpha(1)-(Gq)-signalling to Erk1/2 activation is not mediated via EGF receptor transactivation. Exp. Cell Res. 319, 2718–2727 (2013).
Gnad, T. et al. Adenosine activates brown adipose tissue and recruits beige adipocytes via A2A receptors. Nature 516, 395–399 (2014).
Sharman, J. L. et al. IUPHAR-DB: updated database content and new features. Nucleic Acids Res. 41, D1083–D1088 (2013).
Schrage, R. et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat. Commun. 6, 10156 (2015).
Conklin, B. R. et al. Engineering GPCR signalling pathways with RASSLs. Nat. Methods 5, 673–678 (2008).
Armbruster, B. N., Li, X., Pausch, M. H., Herlitze, S. & Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl Acad. Sci. USA 104, 5163–5168 (2007).
D'Orleans-Juste, P., Plante, M., Honore, J. C., Carrier, E. & Labonte, J. Synthesis and degradation of endothelin-1. Can. J. Physiol. Pharmacol. 81, 503–510 (2003).
Uchida, Y. et al. Endothelin-1, but not endothelin-3, suppresses lipoprotein lipase gene expression in brown adipocytes differentiated in culture. Eur. J. Pharmacol. 291, 33–41 (1995).
Mitchell, J. R. et al. Regulation of expression of the lipoprotein lipase gene in brown adipose tissue. Am. J. Physiol. 263, E500–E506 (1992).
Haas, B. et al. Protein kinase g controls brown fat cell differentiation and mitochondrial biogenesis. Sci. Signal. 2, ra78 (2009).
McDonald, M. E. et al. Myocardin-related transcription factor A regulates conversion of progenitors to beige adipocytes. Cell 160, 105–118 (2015).
Shimizu, I. et al. Vascular rarefaction mediates whitening of brown fat in obesity. J. Clin. Invest. 124, 2099–2112 (2014).
Amisten, S. et al. An atlas of G-protein coupled receptor expression and function in human subcutaneous adipose tissue. Pharmacol. Ther. 146, 61–93 (2015).
Regard, J. B., Sato, I. T. & Coughlin, S. R. Anatomical profiling of G protein-coupled receptor expression. Cell 135, 561–571 (2008).
Xue, R. et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat. Med. 21, 760–768 (2015).
Pernow, J., Shemyakin, A. & Bohm, F. New perspectives on endothelin-1 in atherosclerosis and diabetes mellitus. Life Sci. 91, 507–516 (2012).
Crissey, J. M. et al. Adipose tissue and vascular phenotypic modulation by voluntary physical activity and dietary restriction in obese insulin-resistant OLETF rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 306, R596–R606 (2014).
Lee, Y. H., Petkova, A. P., Mottillo, E. P. & Granneman, J. G. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab. 15, 480–491 (2012).
Li, Y. Q. et al. Gq/11alpha and Gsalpha mediate distinct physiological responses to central melanocortins. J. Clin. Invest. 126, 40–49 (2015).
Jennissen, K. et al. A VASP-Rac-soluble guanylyl cyclase pathway controls cGMP production in adipocytes. Sci. Signal. 5, ra62 (2012).
Chen, Y. et al. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat. Commun. 4, 1769 (2013).
Elabd, C. et al. Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem Cells 27, 2753–2760 (2009).
Hoffmann, L. S. et al. Stimulation of soluble guanylyl cyclase protects against obesity by recruiting brown adipose tissue. Nat. Commun. 6, 7235 (2015).
Betz, M. J. et al. Presence of brown adipocytes in retroperitoneal fat from patients with benign adrenal tumors: relationship with outdoor temperature. J. Clin. Endocrinol. Metab. 98, 4097–4104 (2013).
Insel, P. A. et al. G protein-coupled receptor (GPCR) expression in native cells: "novel" endoGPCRs as physiologic regulators and therapeutic targets. Mol. Pharmacol. 88, 181–187 (2015).
Kalinec, G., Nazarali, A. J., Hermouet, S., Xu, N. & Gutkind, J. S. Mutated alpha subunit of the Gq protein induces malignant transformation in NIH 3T3 cells. Mol. Cell. Biol. 12, 4687–4693 (1992).
Schroder, R. et al. Deconvolution of complex G protein-coupled receptor signalling in live cells using dynamic mass redistribution measurements. Nat. Biotechnol. 28, 943–949 (2010).
Pfeifer, A. Lentiviral transgenesis. Transgenic. Res. 13, 513–522 (2004).
Kloting, N. et al. Insulin-sensitive obesity. Am. J. Physiol. Endocrinol. Metab. 299, E506–E515 (2010).
Acknowledgements
We thank S. Kipschull and P. Zehner for expert technical assistance and Jennifer Naumann for assistance with culturing and differentiation of human cells. We thank C. Dani (University of Nice Sophia Antipolis) for providing hMADS, S. Gutkind and B. Roth for providing GqQL and Dq constructs, respectively. We thank the DFG for financial support (especially the FOR2372 and GRK1873).
Author information
Authors and Affiliations
Contributions
K.K. designed and performed most experiments, analysed the data and wrote the manuscript. A.K. planned experiments. T.G. analysed the experiments and wrote the manuscript. B.H. performed DMR experiments. A.W. performed GPCR array experiments and analysed the data. A.B. helped with in vivo experiments. A.G. helped with in vivo experiments. L.M.B. analysed GPCR array data and helped write the manuscript. K.S. performed the IP3 experiments and helped with the DMR experiments. M.E.L. performed qPCR and analysed human PCR data. M.J.B. collected human BAT samples. S.E. analysed human PCR data. J.W. planned experiments. M.F. helped generating transgenic mice. M.B. provided human WAT data. G.K. supplied FR. E.K. planned experiments and analysed DMR data. P.I. planned GPCR screen and analysed data and wrote manuscript. A.P. supervised experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-4 and Supplementary Tables 1-3 (PDF 1223 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Klepac, K., Kilić, A., Gnad, T. et al. The Gq signalling pathway inhibits brown and beige adipose tissue. Nat Commun 7, 10895 (2016). https://doi.org/10.1038/ncomms10895
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms10895
This article is cited by
-
Purinergic receptor: a crucial regulator of adipose tissue functions
Purinergic Signalling (2023)
-
The Bacterial Gq Signal Transduction Inhibitor FR900359 Impairs Soil-Associated Nematodes
Journal of Chemical Ecology (2023)
-
An update on the secretory functions of brown, white, and beige adipose tissue: Towards therapeutic applications
Reviews in Endocrine and Metabolic Disorders (2023)
-
EBI2 is a negative modulator of brown adipose tissue energy expenditure in mice and human brown adipocytes
Communications Biology (2022)
-
SIRT7 suppresses energy expenditure and thermogenesis by regulating brown adipose tissue functions in mice
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.