Abstract
Gap junction channels mediate intercellular signalling that is crucial in tissue development, homeostasis and pathologic states such as cardiac arrhythmias, cancer and trauma. To explore the mechanism by which Ca2+ blocks intercellular communication during tissue injury, we determined the X-ray crystal structures of the human Cx26 gap junction channel with and without bound Ca2+. The two structures were nearly identical, ruling out both a large-scale structural change and a local steric constriction of the pore. Ca2+ coordination sites reside at the interfaces between adjacent subunits, near the entrance to the extracellular gap, where local, side chain conformational rearrangements enable Ca2+chelation. Computational analysis revealed that Ca2+-binding generates a positive electrostatic barrier that substantially inhibits permeation of cations such as K+ into the pore. Our results provide structural evidence for a unique mechanism of channel regulation: ionic conduction block via an electrostatic barrier rather than steric occlusion of the channel pore.
Similar content being viewed by others
Introduction
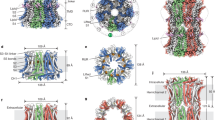
Connexins (Cx) are a 21-member family of integral membrane proteins with four transmembrane (TM) domains (M1–M4), two extracellular loops (E1 and E2), a cytoplasmic (CL) M2–M3 loop and cytoplasmic N- and C-terminal domains (NT and CT; Fig. 1a). Cx monomers assemble as hexameric hemichannels (or connexons) with a central axial pore. Hemichannels from adjacent cells dock to form intercellular, dodecameric gap junction channels (GJCs; Fig. 1b), which can cluster to form gap junction plaques in the plasma membrane (Fig. 1c)1,2. These dense, quasi-hexagonal arrays enabled electron microscopy of isolated plaques and low-resolution, three-dimensional, image reconstruction3. Electron cryomicroscopy (cryoEM) of GJCs assembled from recombinant, human Cx43 at 7.5 (ref. 4) and 5.7 (ref. 5) Å in-plane resolution showed that each hemichannel is comprised of 24 TM α-helices, and the pore has a minimum diameter of ∼15 Å. The extracellular vestibule of GJCs is bounded by a wall of protein, forming a tight seal that excludes extracellular ions and small molecules. An X-ray structure of dodecameric Cx26 at 3.5 Å resolution (Protein Data Bank (PDB) code 2ZW3) revealed that M1 and M2 form the lining of the pore6.
(a) All connexins share a similar topology, with four transmembrane α-helices (M1–M4), two extracellular loops (E1 and E2) and three cytoplasmic domains: the N-terminal tail (NT), M2–M3 cytoplasmic loop (CL) and the carboxy-tail (CT). Over 100 point and deletion mutants have been identified in Cx26 that are linked to deafness. Sixty-nine of these mutants have been tested for trafficking and functionality and are mapped onto the Cx26 topology diagram, indicated by circles coloured solid black with white lettering. Shown at the upper left are the specific residues from adjacent subunits (E42 in subunit B and E47 and G45 in subunit A) identified in this study that coordinate Ca2+ (yellow circle). (b) The gap junction channel (cyan) as derived from the Ca2+-bound crystal structure reported here is comprised of two hexameric Cx hemichannels. The hemichannels dock end-to-end to form an intercellular channel that spans the membrane bilayers of apposed cells and the extracellular gap, from which the name is derived. The phosphorous atoms (red spheres) in the lipid head groups delimit the membrane bilayer with internal aliphatic lipid chains (gold). The ions, depicted as spheres sized by their relative ionic radii, are K+ (green), Cl− (purple) and Na+ (orange). As revealed in this study, the Ca2+ ions (yellow) bind to residues at the interface between the membrane and extracellular gap. The NT, CL and CT of each Cx subunit are not shown, as their densities were not well defined in the map. (c) Representation of a gap junction plaque comprised of channels that pack with quasi-hexagonal symmetry. The cross-section at the right enables visualization of the intercellular pore and the location of bound Ca2+ ions at the interface between the transmembrane α-helices and the extracellular gap. Protein, ions and the membrane bilayers are colored as in b.
GJCs enable direct cell-to-cell exchange of hydrated ions, second messengers and metabolites up to ∼14 Å in the minor dimension7,8, a function necessary for cell differentiation, growth, cell synchronization and coordinated metabolism. For example, GJCs make the heart a functional syncytium, mediating action potential propagation and ionic conduction between cardiac myocytes, thereby regulating the normal heartbeat and coordinated contraction of the muscle, but also mediating potentially fatal cardiac arrhythmias9. GJCs are also responsible for homo- and hetero-cellular communication in vascular endothelial and smooth muscle cells, enabling orchestrated cellular activity over long distances in response to rapidly changing tissue demands10.
Owing to their diverse and specialized functions, Cx isoforms have highly variable selectivities, including moderate to significant charge preferences and distinct permeabilities to signalling molecules such as ATP and inositol triphosphate (IP3), with a molecular weight (MW) cutoff of about 1,000 Da and a size limit of up to ∼14 Å (refs 11, 12). Many of the processes involving GJCs and hemichannels require proper and tightly controlled regulation, as even modest changes in channel activity or permeability can have a large influence on intercellular signalling and cell viability. A variety of physiological processes and pathological states affect channel activity, including pH8,13, phosphorylation10, membrane lipids14, membrane-active agents15, membrane potential16, trans-junctional voltage (Vj)17, loop-gating18,19 and intracellular divalent cations20 including Ca2+ (ref. 21). In some cases, regulation by intracellular Ca2+ may involve calmodulin22.
In the setting of tissue injury, exemplified by myocardial infarction and stroke, cellular ischaemia impairs membrane integrity, resulting in release of Ca2+ from intracellular stores such as the sarcoplasmic reticulum. Ca2+ overload during myocardial ischaemia is characterized by a variety of pathological manifestations, including contractile dysfunction, exacerbation of tissue injury and heart failure23,24. Amelioration of this process represents a potential therapeutic goal for cardiac and cerebral tissue protection during myocardial infarction and stroke. Under normal physiological conditions, ventricular myocytes are electrically coupled by GJCs. Ca2+ overload closes GJCs, which likely contributes to the electrical instability and arrhythmias that occur during myocardial ischaemia.
This structural study is focused on the effects of Ca2+ on Cx26 (Fig. 1a), which forms homomeric and heteromeric channels (for example, with Cx30 or Cx32) that are permeable to cyclic AMP, IP3, glucose and ions with a moderate selectivity for cations11,12,25. Cx26 is involved in hepatic, neural, respiratory, secretory gland, cutaneous and inner ear systems. There are two cochlear gap junction systems (epithelial and connective tissue) in which Cx26 GJCs contribute to K+ recycling and glucose homeostasis26,27. Over 100 Cx26 point mutations have been identified that result in syndromic and non-syndromic deafness in humans. A cluster of deafness-causing mutations is located at the M1/E1 boundary28 (Fig. 1a), which coincides with the location of Ca2+ coordinating residues identified in this study. Previous experimental studies involving multiple Cx isoforms reveal this segment as uniquely important for loop-gating29,30,31 and regulation by divalent metals20.
Low-resolution X-ray scattering32, negative-stain electron microscopy33 and cryoEM and image analysis3 of isolated liver gap junction plaques suggested that Ca2+ elicits large-scale conformational changes. Viewed along the channel axis, the sliding and tilting of Cx subunits within each hemichannel was likened to the closure of a camera iris. Indeed, this has been the prevailing model for Ca2+-induced GJC gating over the last three decades. Projection images along the pore axis of hemichannels by atomic force microscopy supported the model34. The 2ZW3 structure6 provided valuable insight into the architecture of the channel but did not reveal a mechanism for regulation by Ca2+.
To explore the mechanism by which Ca2+ blocks intercellular communication during tissue injury, we determined the X-ray crystal structures of the human Cx26 GJC with and without bound Ca2+. Strikingly, the Ca2+-bound and Ca2+-free channels are nearly identical, ruling out a large-scale steric mechanism for channel block, as well as a local constriction of the pore. Instead, computational analysis suggests that the basis for Ca2+-mediated cation exclusion in GJCs is electrostatic in nature and not due to physical occlusion of the pore.
Results
Determination of Cx26 GJC X-ray structures
Human Cx26 was expressed in Sf9 insect cells and purified from enriched membrane preparations (Supplementary Fig. 1). Crystals that displayed isotropic diffraction were grown in the presence or absence of 20 mM Ca2+ using a novel facial amphiphile detergent (FA-3)35. Both forms crystallized in the H32 space group with two monomers in the asymmetric unit (AU; Fig. 2a). Crystals grown with and without Ca2+ diffracted to resolutions of 3.3 and 3.8 Å, respectively. We solved the Ca2+-bound Cx26 structure independently of the existing Cx26 structure (PDB ID 2ZW3); (ref. 6; see Methods and Supplementary Discussion for details). Molecular replacement (MR) with a dimer of a Cx Cα model based on our cryoEM map (PDB ID 1TXH5) resulted in X-ray crystallographic maps of sufficient quality to determine the helix directions and refine the relative orientations of the monomers. A new, extended polyalanine MR model was then built based on the 5.7 in-plane × 19.8 Å cryoEM map from two-dimensional crystals of intact Cx43 GJCs5 (Fig. 2b). MR with the hybrid cryoEM/X-ray dimer model and the X-ray crystallographic data resulted in improved initial phases that enabled final model building and refinement (see Methods for details).
(a) Packing of Cx26 dodecamers (cyan) in the H32 lattice. The main crystal contact involves residues on the cytoplasmic surface of the channel. The two subunits in the asymmetric unit, a dimer of Cx26, are shown in green and orange. (b) Three-dimensional cryoEM map at 5.7 Å resolution in X, Y and 19.8 Å resolution in Z derived from electron cryocrystallography of two-dimensional crystals of human Cx43 channels in gap junction plaques5. (c) Ca2+-bound Cx26 structure (cyan) and the 2Fo−Fc electron density map (blue mesh) at 3.3 Å resolution and contoured at 1.5-σ. (d) Side view of the dodecameric assembly of Cx26 forming the GJC. Ca2+ atoms are shown as yellow spheres. Distances are measured between Cα atoms. (e) A cross-sectional view of the Ca2+-bound structure (cyan) and map (blue mesh) showing the funnel-shaped cytoplasmic region of the pore that narrows in proceeding to the extracellular vestibule. (f) Inter-subunit residues that form the Ca2+-binding sites, as revealed by the 3.3 Å resolution crystal structure. The difference density peaks (the Fo−Fc maps) indicate the unrefined positions of Ca2+ ions (green). Three binding sites are shown in this cytoplasmic view. (g) A close-up of one of the Ca2+-binding sites shown in f. (h) A cross-sectional view of the Ca2+-free structure (orange), shown with a 3.8 Å resolution 2Fo–Fc map (blue mesh). (i) A view of the Ca2+-free structure as shown in f for the Ca2+-bound structure, showing both 2Fo−Fc and Fo−Fc maps. Note the absence of any green difference density peaks in the Fo–Fc map. (j) A close-up of E47 in subunit A for the Ca2+-free structure. To better visualize the conformational change of E47, the molecule has been rotated significantly compared with the perspectives shown in g and i. Side chains are shown as sticks, with atoms coloured as: carbon, cyan (Ca2+ bound) or orange (Ca2+ free); oxygen, red; nitrogen, blue. The 2Fo−Fc maps (blue mesh) and the Fo–Fc maps (green) are contoured at 1.5-σ and 4-σ, respectively.
Cx26 crystallized as a dodecameric channel, which is aligned along two crystallographic axes. The sixfold axis of the hemichannel is aligned along the crystallographic threefold axis, and the docking interface between hemichannels is oriented along the crystallographic twofold axis. The crystals exhibit type II packing, with the dodecameric channels packed in layers that are separated by approximately the dimensions of one hemichannel (abc=75 × 75 × 40 Å3; Fig. 2a). There is a single unique crystal contact between dodecamers in adjacent lattice layers. Aided by prime-and-switch phase improvement36, nearly all of the amino-acid side chains of the TM and EC domains were eventually accounted for in the electron density maps (Fig. 2c–e). The Ca2+-bound Cx26 structure was built and ultimately refined to 3.3 Å resolution (R/Rfree=0.280/0.313). Although the maps are generally of high quality for the TM α-helices and the extracellular loops, the NT, CL and CT domains could not be modelled because the electron density in those regions is not well defined. The final model consists of residues 19–95 and 134–216 for protomer A and residues 21–95 and 134–213 for protomer B. The Ca2+-bound Cx26 structure was used as a search model for MR to solve the structure of Ca2+-free Cx26 at 3.8 Å resolution (R/Rfree=0.301/0.333; Fig. 2h). Crystallographic data, structure refinement and validation statistics are shown in Supplementary Table 1. Although the structures possess superb refinement and validation statistics with reasonable stereochemistry, interpretation of models based on maps at modest resolutions, as is the case here, should be approached with caution.
Analytical size exclusion chromatography (SEC) and multi-angle light scattering indicate that Cx26 purified in FA-3 is a dodecamer (Supplementary Fig. 1a,d), forming a thermally stable (Supplementary Fig. 1b) and homogenous population of defined oligomers (Supplementary Fig. 1c). In addition, there is close similarity between the Cx43 GJC cryoEM map (Fig. 2b) and the X-ray structures (Fig. 2c,e,h). These observations demonstrate that Cx26 crystallized as a bona fide GJC.
Ca2+-bound and Ca2+-free structures recapitulate the fold of 2ZW3
Although the maximum RMSDs between our structures and 2ZW3 are substantial (Supplementary Fig. 2 and Supplementary Table 2), our independently determined structures recapitulate the general fold of 2ZW3 (ref. 6 and Supplementary Fig. 2). The channel dimensions of the present models are also similar to those reported previously for 2ZW3 (ref. 6): 120 Å from the cytoplasm to the cytoplasm with the pore diameters at the cytoplasmic entrance and the docking interface of 50 and 25 Å, respectively (Fig. 2d). In addition, the docking interactions between hemichannels are very similar in our structures and 2ZW3, and include hydrogen bonding between Q57 residues and between N176 and D179 of apposed hemichannels. The role of the interactions between N176 and D179 in maintaining junctional integrity was recently demonstrated37, and is consistent with experiments on docking of compatible Cx isoforms38.
Ca2+ is coordinated by adjacent subunits
In the presence of Ca2+, Fo–Fc difference density maps display two discrete 5 σ peaks in the AU between subunits at the M1/E1 boundary (green in Fig. 2f,g), corresponding to 6 peaks per hemichannel and 12 peaks per GJC. The difference density peaks reside between the carboxylates of two glutamate residues and the carbonyl oxygen of a glycine, suggestive of a divalent cation-binding site. Ca2+ ions were modelled into the difference density peaks, and the coordination distances were consistent with Ca2+ binding. The average coordination distance in the refined model (2.6 Å), the coordination number (CN=5) and the formal charge of the site (-2) are consistent with mean values reported in an analysis of nearly 1,500 non-EF hand Ca2+-binding proteins for which high-resolution crystal structures were available39.
Comparison of the Ca2+-bound and Ca2+-free electron density maps corroborates the identity of the Ca2+ ions: the peaks ascribed to Ca2+ in the Ca2+-bound Cx26 structure were absent in the Ca2+-free Fo–Fc maps (Fig. 2i,j). The identity of Ca2+ ions is therefore supported by (i) the geometries and distances to coordinating oxygen atoms, (ii) the Fo–Fc (Fig. 2f,g) and Fo–Fc omit maps, (iii) anomalous difference maps from the Ca2+-bound crystals (Supplementary Fig. 3) and (iv) the absence of difference density peaks in the Ca2+-free maps (Fig. 2i,j; see Methods for details). Given the identification of the difference peaks as arising from bound Ca2+, the sites and Ca2+ coordinating residues are shown in Fig. 3a–d. The configuration and interactions of these residues in the Ca2+-free structure are shown in Fig. 3e–h.
The Ca2+-bound (cyan) and Ca2+-free (orange) structures, as well as the Ca2+ ions (yellow) are coloured identically to Fig. 2. (a,e) Cytoplasmic, axial view of the (a) Ca2+-bound and (e) Ca2+-free hemichannel, with three subunits (A, B and C) showing labels for TM helices M1–M4 in subunit A. (b,f) Close-up, cytoplasmic views of Ca2+ coordination sites in the (b) Ca2+-bound and (f) Ca2+-free structures. The dashed boxes delimit the residues involved in coordination of a single Ca2+ ion. (b) and their interactions in the absence of Ca2+ (f). The Ca2+ atoms are bound between adjacent subunits, utilizing a pentamer of protein ligands, and exhibit hemispheric coordination, with the unliganded face exposed to bulk solvent in the pore of the channel. (c,g) Side views of the (c) Ca2+-bound and (g) Ca2+-free hemichannels. For clarity, the three subunits nearest to the viewer have been removed. (d,h) Close-up, side views of the Ca2+ coordination sites in the (d) Ca2+-bound and (h) Ca2+-free structures. The dashed boxes delimit the residues involved in coordination of a single Ca2+ ion (d) and their interactions in the absence of Ca2+ (h). Ca2+ coordination and hydrogen bonding are shown as solid black lines.
Ca2+ ions in adjacent binding sites are separated by 16 Å, with a 60° rotation around the pore axis. The maximum distance between the Ca2+-binding sites across the pore lumen is 23 Å. The Ca2+-binding sites coincide with the region of closest approach between the pore-lining M1 helices of adjacent subunits in the hemichannel. However, the major inter-subunit interface is between M1 and M2 (> 500 Å2), and includes a salt bridge between E42 and R75, which is not present in the Ca2+-free structure, as discussed below. The proximity of the adjacent M1 helices in this region and the substantial M1–M2 contact surface are both conserved in the presence or absence of Ca2+ binding (Fig. 3a,e), as well as in 2ZW3 (ref. 6).
Ca2+ coordination is mediated by the carboxylate of E47 and the carbonyl oxygen of G45 in one subunit and the carboxylate of E42 in the adjacent subunit (Fig. 3b,d). Thus, it is not surprising that previous sequence-based analyses did not predict this Ca2+-binding site as the quaternary structure of the site involves residues in adjacent subunits. The five oxygen atoms that serve as the protein ligands for coordination of the Ca2+ ions adopt a square pyramidal geometry, common with CN=5 (ref. 39). Hemispheric coordination of Ca2+ leaves the bound ion exposed to the aqueous channel (Fig. 3b), where the Ca2+ is expected to remain partially hydrated (as shown by molecular dynamics (MD) simulations discussed below). The carboxylates of the glutamate residues act as coordinating bidentate ligands, with equivalent Ca2+ coordination distances and angles between the Oɛ1 and Oɛ2 oxygens.
The Ca2+-bound and Ca2+-free structures are nearly identical
Unlike most ion channels, atomic ions are not dehydrated as they pass through GJCs40. The pores of the Ca2+-bound and Ca2+-free channel structures are nearly identical, with a limiting diameter of ∼15 Å (measured between K41 side chains of opposing subunits; Fig. 4a–c and Supplementary Fig. 2d,e). Thus, either channel can easily accommodate fully hydrated K+ or Cl− ions as both ions have a first hydration shell diameter of 6.6 Å (Fig. 4c,d). In addition, the Ca2+-bound and Ca2+-free structures are nearly identical (average Cα RMSD=0.4 Å; Fig. 4a,c). Thus, no major subunit rearrangements are associated with Ca2+ binding, and physical occlusion of the pore is not the cause of Ca2+-dependent effects on GJC permeability.
The Ca2+-bound (cyan) and Ca2+-free (orange) structures, as well as the Ca2+ ions (yellow) are coloured identically to Figs 2 and 3. (a) Side views of superimposed Ca2+-bound and Ca2+-free GJC structures. (b) Plots of distance from the centre of the extracellular gap versus pore radius for the Ca2+-bound channel (cyan line) and the Ca2+-free channel (orange line). The solid horizontal lines at ±28 Å indicate the locations of the Ca2+-binding sites within each hemichannel. (c) Overlay of the Ca2+-bound (cyan) and Ca2+-free (orange) hemichannels, viewed from the extracellular gap and shown at the level of the minimum pore diameter (∼15 Å, as measured between Nζ atoms of K41) within the hemichannel, located between K41 residues of subunits that are directly across from one another. The side chains for the K41 residues are shown as sticks. A K+ ion is shown at the centre of the hemichannel with its first (7 Å diameter) and second (12 Å diameter) hydration shells shown in dark and light blue, respectively. (d) A van der Waals surface representation of the Ca2+-free Cx26 hemichannel. The K+ ion and hydration spheres are shown as in c.
Conformational changes are localized at the Ca2+-binding sites
The most significant structural differences between the Ca2+-bound and Ca2+-free channels occur at the Ca2+-binding sites (Fig. 5a–e; Supplementary Fig. 4 and Supplementary Movies 1,2). In the absence of Ca2+, the E47 side chain swings by >90o and 7 Å away from the Ca2+ coordinating conformation (measured at the Cδ atom) to form an intrasubunit salt bridge with K188, the position of which is essentially invariant between the two structures (compare Fig. 5a,c with b,d). E42 also adopts an alternate conformation in the Ca2+-free structure. The coordinating carboxylate of E42 in the Ca2+-bound structure rotates ∼90° and forms an intrasubunit hydrogen bond with the Nɛ of R75 (Supplementary Fig. 4). The carbonyl of G45 moves ∼1 Å closer to E42, resulting in an additional intersubunit van der Waals contact in the absence of Ca2+. The protein–protein interactions made by E47, E42 and G45 in the Ca2+-free structure result in a modest main chain rearrangement relative to the Ca2+-bound structure. Specifically, the Ca2+-free backbone between residues W44 and Q48 shifts an average of 0.8 Å away from the pore centre and slightly towards M1 of the adjacent subunit.
The Ca2+-bound (cyan) and Ca2+-free (orange) structures, as well as the Ca2+ ions (yellow) are coloured identically to Figs 2, 3, 4. (a,c) Close-up cytoplasmic and side views, respectively, of the Ca2+ site in the Ca2+-bound structure, corresponding to the dashed boxes in b and d of Fig. 3. (b,d) Close-up cytoplasmic and side views, respectively, of the Ca2+ site in the Ca2+-free structure, corresponding to the dashed boxes in f and h of Fig. 3. (e) Superposition of the Ca2+-free structure (orange) onto the Ca2+-bound structure (cyan). The E47 side chain adopts different rotamer configurations in the two structures, with the carboxylate stabilized by an ionic interaction with the terminal amine of the K188 side chain in the Ca2+-free structure. The E47 side chain adopts different rotamer configurations in the two structures, with the carboxylate stabilized by an ionic interaction with the terminal amine of the K188 side chain in the Ca2+-free structure (b,d). (f) Sequence propensity diagram of all 21 human Cx isoforms spanning residues 40–50 that include the Cx26 Ca2+-binding residues; this is also a locus harbouring several Cx26 deafness mutations (A40, W44, G45, D46, E47 and D50). Residue numbering is based on the Cx26 sequence. Yellow circles indicate the positions of the residues that coordinate Ca2+, and deafness-causing mutations are shown below the residue number.
Among Cx isoforms, most of the native residues are highly conserved in the vicinity of the Ca2+-binding site (residues 40–50; Fig. 5f). An acidic residue is conserved in all Cx isoforms at position 47 (Cx26 numbering), whereas an acidic residue at position 42 is absent in 15 of 21 Cx isoforms. However, 11 of these 15 isoforms have an acidic residue (glutamate) at position 41. Therefore, with just a one residue shift in the primary sequence, it is reasonable to speculate that an intersubunit E41/42(B)-G45(A)-E/D47(A) Ca2+-binding site is a feature of most GJCs.
Electrostatic mechanism for Ca2+-based regulation of permeation
To investigate whether Ca2+ binding significantly alters the electrostatic profile of the channel pore, we calculated surface potentials with (Fig. 6a) and without (Fig. 6b) the contribution of the bound Ca2+ ions. For the Ca2+-bound structure, the five-coordinate Ca2+ ions resulted in an almost entirely positive electrostatic surface potential for the protein lining the pore (Fig. 6a). The Ca2+-free pore had a distinctly different electrostatic landscape, characterized by a variegated electrostatic surface with a strongly negative potential in the region of the acidic Ca2+-binding residues (Fig. 6b). The significant difference between the Ca2+-bound and Ca2+-free surface potentials suggests an electrostatic mechanism by which GJCs can regulate ion permeation. The substantial positive electrostatic surface potential in the Ca2+-bound structure is particularly informative, as we did not observe physical closure of the channel upon Ca2+ binding.
Depictions of GJC electrostatic surfaces (a,b) and molecular dynamics (MD) simulations (c,d). Comparison of Ca2+-bound (a,c) and Ca2+-free (b,d) states suggests that Ca2+ binding creates a positive surface potential in the pore (blue) that restricts cation permeation. Electrostatic potential surfaces (a,b) with positive and negative electrostatic potentials shown in blue and red, respectively (colour scale is −15 to +15 kT e−1). The protein interior is grey. Cross-sectional side views for (a) Ca2+-bound and (b) Ca2+-free GJCs, coloured according to electrostatic potential. (c,d) Superposition of K+ ion positions (green dots) every 0.1 ns for a 30-ns continuous segment within a 50-ns all-atom MD simulation. Ions are coloured as in Fig. 1b. Cl− ions are not shown for clarity, but a notable observation of the MD simulations is an enhanced occupancy of Cl− ions in the Ca2+-bound pore compared with the Ca2+-free pore (Supplementary Movie 3). (e,f) Hemispheric coordination of Ca2+ ions by the carboxylate of E47 and the carbonyl oxygen of G45 in one subunit and the carboxylate of E42 in the adjacent subunit enables the binding of water molecules on the luminal side of the channel. Two to three water molecules are bound to each Ca2+ ion throughout the MD simulation. The water molecules are shown at a single MD time step.
To determine whether the electrostatic surface potentials affect ion permeation and selectivity, we performed MD simulations of the Ca2+-bound (Fig. 6c) and Ca2+-free (Fig. 6d) X-ray structures embedded in two 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayers. Each system included explicit water molecules and was neutralized and ionized with 140 mM KCl in the cytoplasmic region and 140 mM NaCl in the extracellular region (Fig. 1b), resulting in a total system size of ∼300,000 atoms. For the purposes of electroneutrality, the Cl− concentration was set to 140 mM in the MD calculations. However, we realize that the physiologic cytoplasmic concentration of Cl− is ∼4 mM, with most of the additional negative charge contributed by cytoplasmic proteins. Simulations consisted of unrestrained equilibration (20 ns) followed by a production phase (at least 30 ns; Supplementary Fig. 5). For this analysis, we defined Ca2+ coordination as a distance of <2.6 Å between a coordinating atom and the Ca2+ ion.
In the Ca2+-bound simulation (Fig. 6c and Supplementary Movie 3), Ca2+ ions remained bound, and coordination by the E42 and E47 carboxylates (E42-Oɛ and E47-Oɛ) was stable (Supplementary Figs 6,7), with either three or four of the carboxylate oxygens coordinating the Ca2+ at all times. However, Ca2+ coordination by the carbonyl oxygen of G45 was variable. Depending on the site, coordination by the carbonyl oxygen was maintained throughout, lost or lost and then restored (Supplementary Movie 4). Participation in Ca2+ coordination by G45-O was stable at only one site during the simulation (Supplementary Movie 5). We suspect the loss of Ca2+ coordination by G45-O in the MD simulations may be due to known limitations in the nonpolarizable force field describing the interactions between divalent cations and carbonyl oxygen atoms41. To assess whether loss of G45-O Ca2+ coordination altered the macroscopic Cx26 channel properties, we repeated the MD simulation with restraints on the distances between the G45-O atoms and the Ca2+ ions (Supplementary Figs 8 and 9), which displayed Ca2+ coordination geometry similar to that of the Ca2+-bound X-ray structure. Specifically, Ca2+ coordination by G45-O and all four E42 and E47 carboxylate oxygens was stable at most sites for the majority of the MD simulation, with occasional excursions by a single carboxylate oxygen, which reduced the number of protein ligands to four. Importantly, MD simulations with restraints on the distance between Ca2+ and G45-O did not alter the cationic permeability results without restraints (compare Fig. 6c and Supplementary Fig. 8b). Throughout the simulations, three or four water molecules participated in ion coordination at each Ca2+-binding site, maintaining a total coordination number of seven to eight (Fig. 6e,f).
A notable result of the simulations was the difference in K+ ion occupancy within the pore between the Ca2+-bound and Ca2+-free channels. For the Ca2+-bound channel (Fig. 6c and Supplementary Movie 3), K+ ions were almost completely excluded from the pore. In the Ca2+-free simulation (Fig. 6d and Supplementary Movie 6), K+ ions rapidly entered the channel and clustered around the Ca2+-binding sites. We calculated the potential of mean force (PMF) for K+ and Cl− along the Ca2+-bound and Ca2+-free channel axes based on ion density distributions in the all-atom simulations. The resultant K+ and Cl− free energy profiles (Supplementary Fig. 10) paired with the electrostatic analysis suggest a novel mechanism for Ca2+-dependent ion selectivity, in which Ca2+ binding functions as an electrostatic switch that dramatically reduces cation permeability.
Discussion
We report here the first structural identification of Ca2+-binding sites in GJCs. The X-ray structures of Cx26 GJC in the presence and absence of Ca2+ revealed that Ca2+ is coordinated at the M1/E1 boundary by residues from adjacent subunits. We expect that the Ca2+-binding sites identified here are likely the same as in undocked, functional Cx26 hemichannels, which are also regulated by Ca2+. Ca2+ binding to the Cx26 GJC did not trigger a global conformational change to a sterically closed state, as observed in low-resolution studies of Cx GJCs3 and hemichannels34. As Cx26 was co-crystallized with Ca2+, we assume that large-scale structural rearrangements were possible and would have occurred before crystallization. However, we observed only local conformational changes in the vicinity of Ca2+ coordination, which did not occlude the pore. Likewise, the side chain conformational changes occur for residues adjacent to the lumen of the pore rather than on the perimeter. Therefore, it is unlikely that the observed conformational changes are due to crystal packing artefacts.
Electrostatic calculations and MD simulations revealed that block of K+ permeation is achieved by the extensive positive surface potential conferred by the bound Ca2+; that is, Ca2+-binding creates an electrostatic barrier that impedes cation entry into the pore. We propose that Ca2+ binding in GJCs acts as an electrostatic switch that dramatically shifts the charge selectivity of the channel, creating a strongly positive environment within the pore that inhibits the ability of cations such as K+ to enter and thereby permeate the pore. Given the sequence conservation of residues that participate in Ca2+ binding, we expect that the electrostatic switch regulating Ca2+-dependent ion selectivity is a general mechanism in the family of Cx junctional channels.
In biomedical terms, this study of junctional channels is relevant to conditions of Ca2+ overload, which occur in tissues subject to injury. It is well known that pathologically elevated cytosolic Ca2+ levels cause inhibition of gap junction coupling, reducing the intercellular propagation of toxic signals from dying/diseased cells. It is generally believed that this occurs at Ca2+ levels above micromolar42,43,44. At lower concentrations, there is evidence that Ca2+ can permeate junctional channels44,45,46. Our structural and computational results suggest that Ca2+ binding within GJCs profoundly alters the character of permeating species. Due to the positive electrostatic potentials within the pore that is induced by Ca2+-binding, we expect that the channels will not only become far less permeable to cations but also any molecules with net positive charge.
There is theoretical and electrophysiological evidence that barriers to the flux of molecules and ions can be explained by purely electrostatic effects, without a requirement for steric occlusion47,48. Our results provide the first structural evidence for a unique mechanism of channel regulation: block of ionic conduction via an electrostatic barrier rather than steric occlusion of the pore.
Methods
Expression, solubilization and purification of human Cx26 for crystallization
Expression of human Cx26 C211S, C218S (in which two cysteine residues in the CT were replaced with serine to prevent non-native disulfide bond formation) with a C-terminal hexahistidine tag was performed by infection of Spodoptera frugiperda (Sf9) insect cells with recombinant baculovirus. Cells were infected at a multiplicity of infection (MOI) of 5 and harvested 48–52 h post infection. All subsequent steps were performed at 4 °C, unless otherwise indicated. Cells were lysed by 20–25 strokes using a Dounce homogenizer in low-salt buffer (50 mM HEPES, pH 7.5, 50 mM NaCl, 0.5 mM EDTA and 1 × Complete protease inhibitor cocktail (Roche)). Nucleic acids were digested by adding 10 units of Benzonase (EMD Biosciences), adjusting the buffer to include 2.5 mM MgCl2, followed by 15 min of gentle rocking. The lysate was clarified by ultracentrifugation (100 k × g, 30 min, Beckman model Optima LE-80K), and the pellet was washed with low-salt buffer, high-salt buffer (low-salt buffer with NaCl increased to 1 M) and final wash buffer (high-salt buffer without EDTA). The final pellet contained enriched plasma and organellar membranes. Membranes were stored as aliquots, flash frozen in liquid N2 and stored at −80 °C until use.
Membranes (∼5 g wet weight) were thawed on ice and resuspended in 50 ml extraction buffer (50 mM HEPES, pH 7.5, 300 mM NaCl, 10 mM imidazole, 2% (v/v) glycerol and 1 × Protease Inhibitor Set V (Calbiochem)). Membranes were disrupted by ten strokes using a Dounce homogenizer in order to obtain a small particle size for efficient solubilization. A 10% (w/v) stock solution of N-decyl-β-D-maltopyranoside (DM) detergent (Anatrace) was added to the resuspended membranes to a final concentration of 1% (w/v; ∼1 mM; ∼10 × critical micelle concentration (CMC)). Membranes were solubilized for 3 h on a rotator and then clarified by ultracentrifugation (150 k × g, 40 min, Beckman model Optima LE-80K). After detergent solubilization, it was found that maintaining the protein in buffers with high ionic strength (0.5–1.0 M NaCl) was essential to ensure protein stability and solubility during affinity purification and subsequent concentration steps. His-tagged Cx26 was purified from solubilized membranes by binding the protein to immobilized cobalt affinity resin (Clontech) at a ratio of 1 ml affinity resin per 50 ml detergent extract. After batch binding for 30–60 min, the slurry was poured into a column, and the beads were washed with at least 5 column volumes (CVs) of extraction buffer with 1% (w/v) DM, followed by 5 CVs of wash buffer (50 mM HEPES, pH 7.5, 1 M NaCl, 0.5% (w/v) DM, 20 mM imidazole and 1 × Protease Inhibitor Set V). DM was exchanged for FA-3 on the column by addition of two CVs of exchange buffer (50 mM HEPES, pH 7.5, 1 M NaCl, 20 mM imidazole, 2.5% (v/v) glycerol and 1 × Protease Inhibitor Set V) containing 0.08% (w/v) FA-3 detergent (4 × CMC) to the resin, and 1 CV was allowed to flow through the column. The column was capped, kept stationary and incubated with the new detergent for 15 min. The resin was further washed with 1 CV of FA-3 exchange buffer. Cx26 was then eluted from the column in 1 ml fractions with elution buffer (exchange buffer containing 250 mM imidazole) containing 0.05% FA-3. Fractions with the highest protein concentrations, as estimated by absorption at A280 (NanoDrop 1000 spectrophotometer; Thermo Scientific), were pooled and concentrated to ∼0.5 ml in a 100-kDa molecular weight cutoff (MWCO) concentrator (Millipore). Imidazole was removed using a G-25 desalting column (GE Healthcare) equilibrated in final buffer (100 mM HEPES, pH 7, 1.0 M NaCl, 2.5% (v/v) glycerol and 1 × Protease Inhibitor Set V) containing 0.02% (w/v) FA-3. For crystallization trials, the eluate from the desalting column (∼1 ml) was concentrated to 4–6 mg ml−1 (150–200 μM) using a 500-μl or 4-ml concentrator (100 kDa MWCO; Millipore).
Crystallization of human Cx26
Crystals of Cx26 in FA-3 could be grown reproducibly at 25 °C using hanging-drop or sitting-drop vapour diffusion methods by mixing 1 ml of Cx26 in final buffer with 0.02% FA-3 and 1 μl of a reservoir composed of 100 mM HEPES, pH 7, 15 mM sodium formate, 20 mM CaCl2 and 12% PEG 3350. Crystals for X-ray diffraction experiments were on average 50 × 50 × 25 μm3 and typically grew within 1–2 weeks in the presence of 5–50 mM CaCl2. The best diffracting Ca2+-bound crystals were grown from setups with 20 mM CaCl2. To grow Ca2+-free Cx26 crystals, the same crystallization setup and reservoir buffer were used, with the CaCl2 omitted from the formulation. In the absence of CaCl2, crystals required at least a month to grow. Cryoprotectant (either glycerol or ethylene glycol) in harvesting buffer (100 mM HEPES, pH 7, 20 mM sodium formate, 25 mM CaCl2 (omitted when harvesting Ca2+-free crystals), 14% PEG3350 and 0.02% FA-3) was added directly to the crystallization drop by increasing the concentration of glycerol or ethylene glycol in the crystallization drop in 5% increments every 5–10 min. Crystals were then harvested directly from the drop into 50–100 μm diameter loops (Hampton Research), flash frozen by plunging into liquid N2 and loaded into ALS-style pucks.
Synchrotron X-ray data collection
For the Ca2+-bound Cx26 crystals, X-ray diffraction data were collected at the SER-CAT 22-ID beam line at the Advanced Photon Source at Argonne National Laboratory. Initial indexing of the crystals by HKL2000 (ref. 49) suggested a rhombohedral Bravais lattice (H3), a maximal diffraction resolution of 3.0 Å and a mosaicity of 0.5°. Using a 50-μm beam and 1° oscillations, several wedges of data were collected from one crystal. Ultimately, three data sets were integrated separately and merged upon scaling, with an overall Rmerge of 7.5% and a resolution cutoff of 3.1 Å.
For the Ca2+-free Cx26 crystals, X-ray diffraction data were collected at the GM/CA-CAT 23-ID beam line at the Advanced Photon Source. The Ca2+-free crystals had the same Bravais lattice and were isomorphous with the Ca2+-bound crystals, although reflections could only be observed to 3.6 Å resolution. Two wedges of data from one crystal were merged upon scaling, with an overall Rmerge of 7.5% and a resolution cutoff of 3.8 Å. Both crystal forms (Ca2+-bound and Ca2+-free) conformed to the H32 space group with two molecules in the AU with a Matthews coefficient (Vm) of 3.7 and a solvent content of 67%. Crystallographic statistics are listed in Supplementary Table 1. Resolution limits for the data sets were determined based on data completeness, I/s, and CC1/2 (ref. 50) in the highest resolution shells. For the Ca2+-free data sets, a strong solvent pattern in the diffraction images at ∼3.5 Å resolution prevented inclusion of data beyond 3.6 Å resolution. Resolution limits for model refinement and validation were determined based on the CC1/2 values and assessment of the quality of the electron density map.
Structure determination
We solved the Ca2+-bound Cx26 X-ray crystallographic structure first using the previously determined Cx26 structure (PDB ID: 2ZW3)6 as the MR search model. We also solved the structure independently using a MR model based on a 5.7 × 19.8 Å2 resolution cryoEM map derived by analysis of two-dimensional crystals of Cx43 (ref. 5). These two structure determination methods yielded the same solution. An explanation of our reasons for solving the structure independently is included in the Supplementary Discussion. The Ca2+-bound structure presented here is the result of MR with the cryoEM model. Using Molrep51 in CCP4 (ref. 52) and Phaser53 in Phenix54, we performed MR searches for multiple copies of a Cx26 monomer composed of just the four TM domains, which consisted of 87 carbon atoms. These searches failed, but a search with a dimer of the cryoEM based Cα model (PDB ID: 1TXH5) succeeded and revealed two monomers in the AU of the H32 crystals. The resultant maps had clear 2Fo–Fc electron density, corresponding to the TM domains of the Cx26 GJC that extended beyond the limits of the 1TXH Cα model. The biological sixfold axis that gives rise to the Cx26 hemichannel was coincident with the crystallographic threefold axis, and the biological twofold axis that gives rise to the GJC (at the interface between hemichannels) was coincident with the crystallographic twofold. We converted the Cα model to polyalanine (polyAla) in Coot55 and performed rigid body refinement in Phenix. Further refinement resulted in electron density maps of sufficient quality in the TM domain to determine the directionality of the TM α-helices. We reversed the direction of the M2 helix in the 1TXH model and generated new monomeric and dimeric models consisting of ideal polyAla α-helices. We extended this model through rounds of manual building in Coot, refinement in Phenix and density modification and non-crystallographic symmetry (NCS) averaging with RESOLVE36. Although new electron density appeared that could be attributed to polypeptide, the maps did not improve sufficiently to allow additional model building.
We then used the cryoEM map5 to construct a new model beginning with a dimer of the 4-TM bundle, represented as polyAla. Pymol56 was used to manually fit the dimeric polyAla model consisting of 174 residues into the cryoEM map. On the basis of secondary structure predictions and the cryoEM density map, we placed two ideal polyAla α-helices and two β-strands in the extracellular region. We then modelled four additional, polyAla α-helices as cytoplasmic extensions of the TM domains, based on the cryoEM map contoured at 1-σ. The final monomeric model based on the cryoEM map consisted of 184 residues. We generated a dimer of the expanded model by application of the NCS operators determined from the dimeric 4-TM polyalanine model that we built with the aid of the initial X-ray crystallographic maps. It was important to use the NCS operators based on the X-ray crystallographic map to generate the dimer because the relative orientations of the Cx subunits differed slightly between the two- and three-dimensional crystals. The expanded dimeric model consisted of 368 residues.
We used Phaser-MR within Phenix53 for MR, with the expanded dimer as the search model and the H32 X-ray crystallographic data as the target. The search included data from 35 to 3.5 Å resolution and succeeded with a top translation function Z-score of 6.9. Rigid body refinement of the MR model in Phenix yielded crystallographic R and Rfree values of 0.511 and 0.526, respectively, and 2Fo–Fc maps clearly showed density for the full dodecameric channel (Fig. 2c).
We used Coot to rebuild the model based on the X-ray crystallographic maps, removing all but two turns of one α-helix from the EC domain because of poor density and disagreement between the model and maps in that region. We also made contiguous α-helices for the TM domains from the helical fragments used for MR and rigid body refinement. After rounds of real space modification of the polyAla model and refinement, followed by prime-and-switch density modification with RESOLVE36, we were able to identify and position aromatic side chains and disulfides, which enabled assignment of the sequence and the TM α-helices. We first identified M2 by electron density that we determined was the sequence HYFPxxH, corresponding to the Cx26 sequence HYFPISH (residues 67–73) leading into M2 from E1 within the extracellular domain. Next, we identified M3 by density for a polypeptide with the sequence FF(long)xxF (where ‘long’ indicates an extended side chain, corresponding to the Cx26 sequence FFRVIF in the centre of M3 (residues 141–146)). Then, we defined M4 by density for a sequence KxxFxxF, corresponding to the Cx26 sequence KTVFTVF (residues 188–194) at the extracellular end of M4, near the end of E2 in the extracellular domain. Finally, we confirmed the identity of M1 by density for a sequence WxxxxFxF, corresponding to the Cx26 sequence WLTVLFIF (residues 24–31) in the cytoplasmic half of M1, near the NT domain. The overall map quality, including that for the extracellular domain, continued to improve as we added side chains. We began building the backbone of E2, connecting M3 and M4. Then, based on strong density for a disulfide (there are three in the EC domain) and nearby recognizable density for a tryptophan and other bulky side chains, we built side chains for the Cx26 sequence MQRLVKCNAWPCP (residues 163–175). We continued adding sequence in this region based on a second disulfide (between C58 and C180) and a neighbouring phenylalanine (F181). During building and refinement, the maps improved significantly with the appearance of a missing stretch of 14 residues (S162–P175) in E2 of the extracellular domain. The new density revealed a β-strand that, with an existing β-strand, formed a β-hairpin at the extracellular gap (extending from L166–F181). Finally, we were able to complete the model of the TM and EC domains, consisting of residues 18–93 and 135–216 in molecule A (156 of 226 total) and 21–96 and 134–213 in molecule B (154 of 226). Therefore, ∼70% of the total sequence is accounted for in the final model.
Throughout building and refinement, several strong positive Fo–Fc difference density peaks were present that were not accounted for by the protein model, exemplified by the peak in Supplementary Fig. 3a (denoted by ‘?’), which may be a detergent molecule. Two NCS-equivalent peaks in the AU that were greater than 5-σ were located at an interface between Cx subunits, at the end of M1, extending into the start of E1. The peaks localized near the carboxylates of a glutamate from one Cx (E42) and the carboxylates of a second glutamate (E47) and the carbonyl oxygen of a glycine (G45) from the adjacent Cx subunit (Fig. 2e–g). The difference density surrounded by four carboxylates and one carbonyl was strongly suggestive of divalent cation coordination. As the crystals had been grown in the presence of 20 mM CaCl2, we modelled Ca2+ ions into the Fo–Fc density and refined the model with and without Ca2+ coordination restraints in Phenix. Even in the absence of the Ca2+ restraints, the model refined well with the added Ca2+ ions, accounting for the Fo–Fc density and with appropriate distances to the coordinating oxygens.
To help verify the presence of Ca2+ at these sites, we merged the data sets from three different crystals grown in CaCl2 (in order to amplify the weak Ca2+ anomalous signal) and calculated an anomalous difference map at 4 Å resolution using Ca2+-free, model phases. The only peak observable at >4-σ in the anomalous map partially overlapped with one of the Fo–Fc difference density peaks (+5-σ) that had guided the placement of a Ca2+ atom (Supplementary Fig. 3). Although not definitive or necessarily coincident, the overlap of anomalous and difference peaks provides some evidence of a metal-binding site. Significantly, after refinement, there was an absence of negative Fo–Fc density at the binding site and even some residual positive density on the side of the coordinated ion exposed to the pore. Removal of Ca2+ ions from the model, subsequent refinement and calculation of omit maps gave rise to the re-appearance of >4-σ Fo–Fc peaks at the site where Ca2+ ions had been modelled. In addition, anomalous difference maps were calculated, and a >4-σ peak was present at one of the Ca2+-binding sites in the AU (Supplementary Fig. 3). After modelling the Ca2+ ions and subsequent refinement, the binding sites were analysed for agreement with the published literature and databases of Ca2+-binding sites39,57. The coordination number (n=5), formal charge of the site (n=−2) and coordination bonding distances (both average side chain carboxylate distance and average main chain carbonyl distance=2.6 Å) were all within the reported ranges for Ca2+ binding. Although the resolution limit of the diffraction data precluded the definitive placement of water molecules, it is suspected that the residual positive density was likely due to the presence of coordinating water molecules. Water coordination is expected given the hemispheric and incomplete coordination of the Ca2+ by the protein. MD simulations of the Ca2+-bound channel also showed hydration of the bound Ca2+ ions (Fig. 6c,d).
Electron density for additional turns of α-helices was present on the cytoplasmic side of the channel, but the density was not sufficiently well defined to enable modelling of the CL or either of the termini. Both the CL and CT have been shown to be involved in GJC regulation58 and to interact with one another under acidic conditions59. In our crystals grown at pH 7, we suspect that the CT is involved in the primary crystal contact, found near the ends of the M4 helices of adjacent dodecamers in the crystal lattice (Fig. 2a). We note that this crystal contact only occurs at every other subunit.
We solved the Ca2+-free structure by MR with the Ca2+-bound structure, after removal of the Ca2+ ions. The Ca2+-free crystals were isomorphous with the Ca2+-bound crystals, and the initial maps after MR and refinement were of good quality for the TM and extracellular domains. Despite the lower resolution limit of the diffraction data (3.8 Å), prime-and-switch NCS averaging and density modification resulted in maps that allowed us to obtain a well-refined model of the Ca2+-free GJC. Significantly, no peaks in either Fo–Fc or anomalous maps appeared in the vicinity of the Ca2+-binding site identified in the Ca2+-bound structure.
MD simulations
We used VMD 1.9.1 (ref. 60) to build systems comprised of full Cx26 GJCs embedded in two POPC bilayers. We constructed each system by hydrating the protein with Solvate 1.0.1 (ref. 61) and then removed the water molecules in the surrounding lipid-facing portion of the protein based on hydrophobicity and the positions of lipid-facing, aromatic amino acids. We used the VMD Membrane Builder plugin to generate lipid bilayers for use with the CHARMM36 force field62. We manually positioned the bilayers relative to the full GJC. Then, we removed lipids overlapping with the protein and within the channel. To generate the water box for the system, we used the VMD plugin Solvate. Finally, we neutralized and ionized the system using the VMD plugin Autoionize. Last, we selectively removed ions from the parts of the system corresponding to the cytoplasmic or extracellular spaces and then repeated neutralization and ionization in order to ionize the cytoplasmic space with 140 mM KCl and the extracellular space with 140 mM NaCl. In the case of the Ca2+-free channel, we used the same method to add intracellular CaCl2, in order to make the total Ca2+ equivalent in the simulations. (No cytoplasmic Ca2+ was added to the simulations of the Ca2+-bound channel.)
We used the CUDA-enabled version of NAMD 2.9 to perform the equilibrium MD simulations of these systems using the CHARMM36 force field for the POPC bilayers62 and the CHARMM27 (ref. 63) force field for all other atoms, employing the CMAP correction for handling dihedral angles in polypeptides. Admittedly, there are inherent limitations in attempting to simulate the behaviour of metals covalently bound to proteins, mainly due to the lack of well-established and accurate parameters to describe protein–metal coordination in any of the available MD force fields41.
For each simulation, minimization and equilibration followed the procedure described at http:// www.ks.uiuc.edu/Training/Tutorials/science/membrane/mem-tutorial.pdf. We first equilibrated the lipid acyl chains, keeping the rest of the system fixed. Next, we restrained the protein in order to first minimize and equilibrate the rest of the atoms of the system. Then, we released the restraints on the protein and equilibrated the entire system. Finally, for all-atom production runs employing Langevin dynamics, we used a constant temperature of 310 K and constant pressure of 1 atm with 2 fs time steps.
The carbonyl oxygen–Ca2+ interaction energy is underestimated in the CHARMM27 force field41. Thus, we repeated the Ca2+-bound simulation with the G45O-Ca2+ distance restrained to 2.35 Å, with a spring constant of 40 kcal mol−1.
PMF values were calculated based on the K+ and Cl− ion density distributions along the Ca2+-bound and Ca2+-free Cx26 GJC pore (the Z axes) during 30 ns of the all-atom MD simulation production phases (Supplementary Fig. 10). Ions (either K+ or Cl−) were counted at each position along the Z axis with a fixed radius equal to the minimum channel radius (7 Å, at K41) and a ΔZ of 5 Å. Ion densities (<ρ(Z)>) were calculated by dividing the total number of ions at each position on Z axis by the corresponding cylindrical volume. PMFs were ultimately calculated by solving the equation,

PMF values were adjusted so that values in bulk solvent were zero64.
Electrostatic surface calculations by Internal Coordinates Mechanics and the REBEL method
Electrostatic surface potentials of the Ca2+-bound and Ca2+-free Cx26 GJCs were calculated by the Rapid Exact-Boundary Electrostatics (REBEL) method65 as implemented in the Molsoft software package, ICM-Pro66. The REBEL calculations employ a model in which the solvent is a continuous high dielectric medium and the protein molecular surface is the boundary between the solvent and the low dielectric protein. The electrostatic energy is calculated for a set of point charges on the protein surface by solving the Poisson equation (-∇(ɛ (r)∇φ (r))=ρ (r)), where ɛ is the dielectric constant, φ is the electric potential and ρ is the surface charge density. REBEL uses the boundary element method to solve the Poisson equation for the model system described above, where the boundary is the triangulated analytical molecular surface as described by Connolly67. A limitation of the boundary element method is that a computationally expensive integral equation solution is required to determine the charge distribution at the boundary. To make the calculations tractable, the molecular surface is divided into discrete elements. REBEL uses a simplified version of the multigrid boundary element discretization method established by Vorobjev and Scheraga68, in which the number of boundary elements is reduced by combining all of the elements associated with each surface atom. Consequently, the number of composite boundary elements is reduced to the number of surface atoms without a significant loss in the precision of the charge density calculation65.
The parameters for the REBEL boundary element electrostatic calculations were as follows. Partial charges for atoms of amino acids were assigned according to the molecular mechanics force-field named Empirical Conformational Energy Program for Peptides (ECEPP)69; glutamate and aspartate were assigned a unitary negative charge and arginine and lysine were assigned a unitary positive charge. The atomic radii used in the electrostatic and surface calculations were: carbon, 1.700 Å; nitrogen, 1.447 Å, nitrogen in nitro groups, 1.763 Å; oxygen in carbonyl, 1.542 Å; oxygen in hydroxyls, 1.500 Å; sulfur in disulfide bonds, 1.491 Å, and sulfur in thiols, 1.812 Å, calcium ion, 1.650 Å. The dielectric constants inside and outside the protein (bulk solvent) were 4 and 78.5, respectively. The patch size for which individual surface elements were assumed to have the same polarized charge surface density in the REBEL boundary element method was set to 0.5 Å.
In the case of Ca2+-bound Cx26, we performed REBEL calculations with and without the explicit inclusion of the waters that were found to complete Ca2+-coordination in the MD simulations, and we found no significant difference in the resultant electrostatic surface potentials.
Electrostatic surface potentials were also calculated using Poisson–Boltzmann equations as implemented in Adaptive Poisson-Boltzmann Solver (APBS). Surfaces calculated in this way are essentially identical to surfaces calculated with the REBEL method implemented in ICM-Pro.
Sequence analysis and alignment for determining residue propensity
Homologous sequences from residues 40–50 (Cx26 numbering) for all 21 human Cx isoforms were defined by a sequence alignment performed with PRALINE70 and were uploaded to the Weblogo online server (http://weblogo.berkeley.edu) to generate the diagram shown in Fig. 5f.
Additional information
Accession codes: Coordinates and structure factor files for Ca2+-bound and Ca2+-free Cx26 have been deposited in with the Protein Data Bank under PDB ID 5ER7 and 5ERA, respectively.
How to cite this article: Bennett, B. C. et al. An electrostatic mechanism for Ca2+-mediated regulation of gap junction channels. Nat. Commun. 7:8770 doi: 10.1038/ncomms9770 (2016).
References
Yeager, M. & Harris, A. L. Gap junction channel structure in the early 21st century: facts and fantasies. Curr. Opin. Cell Biol. 19, 521–528 (2007).
Yeager, M. Gap Junction Channel Structure in Connexins: A Guide eds Harris A. L., Locke D. 27–75Humana-Springer (2009).
Unwin, P. N. & Ennis, P. D. Two configurations of a channel-forming membrane protein. Nature 307, 609–613 (1984).
Unger, V. M., Kumar, N. M., Gilula, N. B. & Yeager, M. Three-dimensional structure of a recombinant gap junction membrane channel. Science 283, 1176–1180 (1999).
Fleishman, S. J., Unger, V. M., Yeager, M. & Ben-Tal, N. A Cα model for the transmembrane α helices of gap junction intercellular channels. Mol. Cell 15, 879–888 (2004).
Maeda, S. et al. Structure of the connexin 26 gap junction channel at 3.5A resolution. Nature 458, 597–602 (2009).
Harris, A. L. Emerging issues of connexin channels: biophysics fills the gap. Quart. Rev. Biophys. 34, 325–472 (2001).
Nielsen, M. S. et al. Gap junctions. Compr. Physiol. 2, 1981–2035 (2012).
Desplantez, T., Dupont, E., Severs, N. J. & Weingart, R. Gap junction channels and cardiac impulse propagation. J. Membr. Biol. 218, 13–28 (2007).
Johnstone, S., Isakson, B. & Locke, D. Biological and biophysical properties of vascular connexin channels. Int. Rev. Cell Mol. Biol. 278, 69–118 (2009).
Harris, A. L. Connexin channel permeability to cytoplasmic molecules. Prog. Biophys. Mol. Biol. 94, 120–143 (2007).
Kanaporis, G., Brink, P. R. & Valiunas, V. Gap junction permeability: selectivity for anionic and cationic probes. Am. J. Physiol. Cell Physiol. 300, C600–C609 (2011).
Spray, D. C., Harris, A. L. & Bennett, M. V. Gap junctional conductance is a simple and sensitive function of intracellular pH. Science 211, 712–715 (1981).
Locke, D. & Harris, A. L. Connexin channels and phospholipids: association and modulation. BMC Biol. 7, 52 (2009).
Srinivas, S. Pharmacology of Connexin Channels in Connexins: A Guide eds Harris A. L., Locke D. 207–224Humana-Springer (2009).
Revilla, A., Bennett, M. V. & Barrio, L. C. Molecular determinants of membrane potential dependence in vertebrate gap junction channels. Proc. Natl Acad. Sci. USA 97, 14760–14765 (2000).
Harris, A. L., Spray, D. C. & Bennett, M. V. Kinetic properties of a voltage-dependent junctional conductance. J. Gen. Physiol. 77, 95–117 (1981).
Trexler, E. B., Bukauskas, F. F., Kronengold, J., Bargiello, T. A. & Verselis, V. K. The first extracellular loop domain is a major determinant of charge selectivity in connexin46 channels. Biophys. J. 79, 3036–3051 (2000).
Bukauskas, F. F., Angele, A. B., Verselis, V. K. & Bennett, M. V. Coupling asymmetry of heterotypic connexin 45/ connexin 43-EGFP gap junctions: properties of fast and slow gating mechanisms. Proc. Natl Acad. Sci. USA 99, 7113–7118 (2002).
Palacios-Prado, N. et al. Molecular determinants of magnesium-dependent synaptic plasticity at electrical synapses formed by connexin36. Nat. Commun. 5, 4667 (2014).
Iwatsuki, N. & Petersen, O. H. Acetylcholine-like effects of intracellular calcium application in pancreatic acinar cells. Nature 268, 147–149 (1977).
Zou, J. et al. Gap junction regulation by calmodulin. FEBS Lett. 588, 1430–1438 (2014).
Noble, D. & Noble, P. J. Late sodium current in the pathophysiology of cardiovascular disease: consequences of sodium-calcium overload. Heart 92, iv1–iv5 (2006).
Kalogeris, T., Baines, C. P., Krenz, M. & Korthuis, R. J. Cell biology of ischemia/reperfusion injury. Int. Rev. Cell Mol. Biol. 298, 229–317 (2012).
Suchyna, T. M. et al. Different ionic selectivities for connexins 26 and 32 produce rectifying gap junction channels. Biophys. J. 77, 2968–2987 (1999).
Kikuchi, T., Adams, J. C., Miyabe, Y., So, E. & Kobayashi, T. Potassium ion recycling pathway via gap junction systems in the mammalian cochlea and its interruption in hereditary nonsyndromic deafness. Med. Electr. Microscopy 33, 51–56 (2000).
Matsunami, T. et al. Gap junctions mediate glucose transport between GLUT1-positive and -negative cells in the spiral limbus of the rat cochlea. Cell Commun. Adhes. 13, 93–102 (2006).
Xu, J. & Nicholson, B. J. The role of connexins in ear and skin physiology - Functional insights from disease-associated mutations. Biochim. Biophys. Acta 1828, 167–178 (2013).
Pfahnl, A. & Dahl, G. Localization of a voltage gate in connexin46 gap junction hemichannels. Biophys. J. 75, 2323–2331 (1998).
Tang, Q., Dowd, T. L., Verselis, V. K. & Bargiello, T. A. Conformational changes in a pore-forming region underlie voltage-dependent ‘loop gating’ of an unapposed connexin hemichannel. J. Gen. Physiol. 133, 555–570 (2009).
Verselis, V. K., Trelles, M. P., Rubinos, C., Bargiello, T. A. & Srinivas, M. Loop gating of connexin hemichannels involves movement of pore-lining residues in the first extracellular loop domain. J. Biol. Chem. 284, 4484–4493 (2009).
Unwin, P. N. & Ennis, P. D. Calcium-mediated changes in gap junction structure: evidence from the low angle X-ray pattern. J. Cell Biol. 97, 1459–1466 (1983).
Unwin, P. N. & Zampighi, G. Structure of the junction between communicating cells. Nature 283, 545–549 (1980).
Müller, D. J., Hand, G. M., Engel, A. & Sosinsky, G. E. Conformational changes in surface structures of isolated connexin 26 gap junctions. EMBO J. 21, 3598–3607 (2002).
Lee, S. C. et al. Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins. Proc. Natl Acad. Sci. USA 110, E1203–E1211 (2013).
Terwilliger, T. C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003).
Nakagawa, S. et al. Asparagine 175 of connexin32 is a critical residue for docking and forming functional heterotypic gap junction channels with connexin26. J. Biol. Chem. 286, 19672–19681 (2011).
Bai, D. & Wang, A. H. Extracellular domains play different roles in gap junction formation and docking compatibility. Biochem. J. 458, 1–10 (2014).
Kirberger, M. et al. Statistical analysis of structural characteristics of protein Ca2+-binding sites. J. Biol. Inorg. Chem. 13, 1169–1181 (2008).
Verselis, V. & Brink, P. R. The gap junction channel. Its aqueous nature as indicated by deuterium oxide effects. Biophys. J. 50, 1003–1007 (1986).
Sousa, S. F., Fernandes, P. A. & Ramos, M. J. Molecular Dynamic Simulations: Difficulties, Solutions and Strategies for Treating Metalloenzymes in Kinetics and Dynamics Vol. 12, eds Paneth P., Dybala-Defratyka A. 299–330Springer (2010).
Dahl, G. & Isenberg, G. Decoupling of heart muscle cells: correlation with increased cytoplasmic calcium activity and with changes of nexus ultrastructure. J. Membr. Biol. 53, 63–75 (1980).
Spray, D. C., Stern, J. H., Harris, A. L. & Bennett, M. V. Gap junctional conductance: comparison of sensitivities to H and Ca ions. Proc. Natl Acad. Sci. USA 79, 441–445 (1982).
Sáez, J. C., Connor, J. A., Spray, D. C. & Bennett, M. V. Hepatocyte gap junctions are permeable to the second messenger, inositol 1,4,5-trisphosphate, and to calcium ions. Proc. Natl Acad. Sci. USA 86, 2708–2712 (1989).
Christ, G. J., Moreno, A. P., Melman, A. & Spray, D. C. Gap junction-mediated intercellular diffusion of Ca2+ in cultured human corporal smooth muscle cells. Am. J. Physiol. 263, C373–C383 (1992).
Sun, J. et al. Cochlear gap junctions coassembled from Cx26 and 30 show faster intercellular Ca2+ signaling than homomeric counterparts. Am. J. Physiol. Cell Physiol. 288, C613–C623 (2005).
Rostovtseva, T. & Colombini, M. VDAC channels mediate and gate the flow of ATP: implications for the regulation of mitochondrial function. Biophys. J. 72, 1954–1962 (1997).
Cheng, M. H., Coalson, R. D. & Tang, P. Molecular dynamics and brownian dynamics investigation of ion permeation and anesthetic halothane effects on a proton-gated ion channel. J. Am. Chem. Soc. 132, 16442–16449 (2010).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, Macromolecular Crystallography Part A 307–326 (1997).
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D66, 22–25 (2010).
Collaborative Computational Project, Number 4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D50, 760–763 (1994).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D66, 486–501 (2010).
Schrodinger, L. The PyMOL Molecular Graphics System, Version 1.3r1 (2010).
Harding, M. M. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr. D Biol. Crystallogr. 62, 678–682 (2006).
Hirst-Jensen, B., Sahoo, P., Kieken, F., Delmar, M. & Sorgen, P. L. Characterization of the pH-dependent interaction between the gap junction protein connexin43 carboxyl terminus and cytoplasmic loop domains. J. Biol. Chem. 282, 5801–5813 (2007).
Locke, D., Kieken, F., Tao, L., Sorgen, P. L. & Harris, A. L. Mechanism for modulation of gating of connexin26-containing channels by taurine. J. Gen. Physiol. 138, 321–339 (2011).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Grubmüller, H. & Groll, V. SOLVATE. http://www.mpibpc.mpg.de/grubmueller/solvate (1996-2010).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Brooks, B. R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009).
Im, W., Seefeld, S. & Roux, B. A grand canonical Monte Carlo-Brownian dynamics algorithm for simulating ion channels. Biophys. J. 79, 788–801 (2000).
Totrov, M. & Abagyan, R. Rapid boundary element solvation electrostatics calculations in folding simulations: successful folding of a 23-residue peptide. Biopolymers 60, 124–133 (2001).
Abagyan, R., Totrov, M. & Kuznetsov, D. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 15, 488–506 (1994).
Connolly, M. Analytical molecular surface calculation. J. Appl. Crystallogr. 16, 548–558 (1983).
Vorobjev, Y. & Scheraga, H. A fast adaptive multigrid boundary element method for macromolecular electrostatic computations in a solvent. J. Comput. Chem. 18, 569–583 (1997).
Némethy, G. et al. Energy parameters in polypeptides.10. Improved geometrical parameters and nonbonded interactions for use in the ECEPP/3 algorithm, with application to proline-containing peptides. J. Phys. Chem. 96, 6472–6484 (1992).
Heringa, J. Two strategies for sequence comparison: profile-preprocessed and secondary structure-induced multiple alignment. Comput. Chem. 23, 341–364 (1999).
Acknowledgements
This work was supported by NIH grants R01 HL048908 (M.Y.), R01 GM098538 (Q.Z.), R01 GM071872 (R.A.) and P50 GM073197 (R.C.S., M.Y. and Q.Z.). The Center for Membrane Biology at the University of Virginia provided infrastructure for this work. X-ray diffraction data sets were collected at the Advanced Photon Source, Argonne National Laboratory using the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beam line and the GM/CA 23-ID beam line (funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104)). We thank the beam line staffs for their guidance and expertise during data collection experiments. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. NAMD was developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign. We also thank Susan Leonhardt, Yuanzi Hua, Barbie Ganser-Pornillos and Peter Kasson for technical assistance and helpful discussions. We gratefully acknowledge Kelly Dryden for performing negative stain electron microscopy experiments and Peter Horanyi for his assistance with the analytical SEC/multi-angle light scattering experiments. M.Y. is especially grateful to Nigel Unwin for 30 years of encouragement on this project.
Author information
Authors and Affiliations
Contributions
M.Y. conceived the project and was responsible for overall project direction and execution. K.A.B. and B.C.B. performed cloning, protein expression, purification and crystallization. Q.Z. synthesized facial amphiphile detergents. R.S. provided experimental guidance in the early stages of the work. K.A.B., B.C.B. and M.D.P. solved the X-ray structures. M.D.P. designed and performed the molecular dynamics simulations. R.A., C.A. and M.D.P. performed electrostatic calculations. B.C.B., M.D.P., W.E.M., A.L.H. and M.Y. wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-10, Supplementary Tables 1-2, Supplementary Discussion and Supplementary References (PDF 2063 kb)
Supplementary Movie 1
Ca2+ coordination sites reside at the interface between adjacent subunits, near the entrance to the extracellular gap, where local conformational rearrangements enable Ca2+-chelation. Each of the 12 Ca2+ binding sites is comprised of the G45-O and the E47 carboxylate from one subunit, and the E42 carboxylate of the adjacent subunit. The Ca2+-bound and Ca2+-free structures were superimposed, and the morph conformations tool in Chimera was used to create a conformational trajectory between the two structures (morph model, cyan; Ca2+ atom, yellow; coordinating atoms, sticks). (MOV 213 kb)
Supplementary Movie 2
A view showing the transition between the Ca2+-bound and Ca2+-free gap junction channels, highlighting the E47:K188 salt bridge in the Ca2+-free structure. Supplementary Movie 1 is shown at an oblique angle to emphasize the large movement of the E47 side chain, which swings > 90o and 7 Å away from the Ca2+ coordinating conformation to form an intersubunit salt bridge with K188 in the Ca2+-free structure. (MOV 268 kb)
Supplementary Movie 3
Molecular dynamics (MD) simulation of a Ca2+-bound gap junction channel shows that the Ca2+ ions induce a strongly positive electrostatic potential that dramatically restricts K+ permeation. Six Ca2+ ions (yellow) bind at the interface between adjacent subunit within the hemichannels that dock end-to-end to form a gap junction channel. All atom MD simulations were performed on the Cx26 X-ray structure (cyan) embedded in two POPC bilayers (phosphorous atoms, red spheres). The cytoplasm and extracellular spaces were ionized with 140 mM KCl (K+, green and Cl-, purple) and 140 mM NaCl (Na+, orange), respectively. (For the purposes of electroneutrality, the Cl-concentration was set to 140 mM. However, we realize that the physiologic cytoplasmic concentration of Cl- is ~4 mM, with most of the additional negative charge contributed by cytoplasmic proteins.) Water molecules are shown as blue lines. (MOV 16404 kb)
Supplementary Movie 4
Ca2+ ions remain stably bound throughout the MD simulation, with minimal movement of the protein backbone. The E47 carboxylate from one subunit and the E42 carboxylate of the adjacent subunit are stably coordinated throughout the MD simulation, as are three water molecules (red and white sticks). However, coordination by G45-O was only maintained at 1 out of 12 sites in the unrestrained simulation. (Binding site residues shown as sticks; O atoms, red.) (MOV 7728 kb)
Supplementary Movie 5
The Ca2+ coordination geometry determined by X-ray crystallography is maintained throughout the MD simulation at one of the binding sites. Coordination by G45-O was maintained at this particular site. Restrained MD simulations also maintained coordination by G45-O at every binding site. The electrostatic properties of the channel were not influenced in either the restrained or unrestrained MD simulations. Stick representations and colors are as in Supplementary Movie 4. (MOV 7859 kb)
Supplementary Movie 6
MD simulation of the Ca2+-free gap junction channel shows that the channel is highly permeable to K+ ions. During the MD simulation, K+ and Cl- ions enter and traverse the channel, and K+ ions cluster in the vicinity of anionic residues at the Ca2+ binding sites. (MOV 17736 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Bennett, B., Purdy, M., Baker, K. et al. An electrostatic mechanism for Ca2+-mediated regulation of gap junction channels. Nat Commun 7, 8770 (2016). https://doi.org/10.1038/ncomms9770
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms9770
This article is cited by
-
The retinal pigment epithelium displays electrical excitability and lateral signal spreading
BMC Biology (2023)
-
A deep convolutional neural network for segmentation of whole-slide pathology images identifies novel tumour cell-perivascular niche interactions that are associated with poor survival in glioblastoma
British Journal of Cancer (2021)
-
Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 Å
Nature Communications (2020)
-
Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation
Communications Biology (2020)
-
Carbon replicas reveal double stranded structure of tight junctions in phase-contrast electron microscopy
Communications Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.