Abstract
γ-Aminobutyric acid type A and glycine receptors (GABAARs, GlyRs) are the major inhibitory neurotransmitter receptors and contribute to many synaptic functions, dysfunctions and human diseases. GABAARs are important drug targets regulated by direct interactions with the scaffolding protein gephyrin. Here we deduce the molecular basis of this interaction by chemical, biophysical and structural studies of the gephyrin–GABAAR α3 complex, revealing that the N-terminal region of the α3 peptide occupies the same binding site as the GlyR β subunit, whereas the C-terminal moiety, which is conserved among all synaptic GABAAR α subunits, engages in unique interactions. Thermodynamic dissections of the gephyrin–receptor interactions identify two residues as primary determinants for gephyrin’s subunit preference. This first structural evidence for the gephyrin-mediated synaptic accumulation of GABAARs offers a framework for future investigations into the regulation of inhibitory synaptic strength and for the development of mechanistically and therapeutically relevant compounds targeting the gephyrin–GABAAR interaction.
Similar content being viewed by others
Introduction
γ-Aminobutyric acid type A receptors (GABAARs) are ligand-gated ion channels, which mediate the majority of fast inhibitory synaptic transmission in the mammalian central nervous system. Deficits in GABAAR-mediated neurotransmission have been implicated in a wide spectrum of disorders of the central nervous system, such as epilepsy, anxiety, mood disorders and neuro-developmental impairments including autism, fragile X syndrome and schizophrenia1. Accordingly, GABAARs are important, validated drug targets and compounds targeting GABAARs have been extensively explored and successfully used clinically as sedatives, anxiolytics and anticonvulsive drugs, narcotics and anaesthetics, anti-spasmodics, anti-epileptics, hypnotic and analgesic drugs.
GABAARs are pentameric hetero-oligomers assembled from seven different subunit classes with the most common receptor combination being two α, two β and a single γ subunit2. The majority of synaptic GABAAR subtypes are localized and accumulated by the scaffolding protein gephyrin. Gephyrin is a central player at inhibitory synapses and, besides the structural role as a receptor scaffold, gephyrin also acts as a platform for additional protein–protein interactions, bringing receptors, cytoskeletal elements and signalling proteins into close spatial proximity3,4,5. Therefore, insights into the molecular basis of GABAAR clustering at synaptic sites might allow the advance of new therapeutic principles in the treatment of GABAAR-related disorders. Gephyrin is composed of an N-terminal domain (GephG, residues 1–181) and a C-terminal domain (GephE, residues 318–736), which are connected by an unstructured linker (residues 182–317). Gephyrin was discovered6 by co-purification with glycine receptors (GlyRs) and found to be responsible for anchoring and accumulating GlyRs at postsynaptic sites, which is accomplished by the simultaneous binding of gephyrin to the GlyR β subunit7,8,9,10 and elements of the cytoskeleton11,12. A number of gene knockout studies have addressed the role of the gephyrin–GABAAR interaction in vivo. Gephyrin knock-out mice die within hours after birth13 and analysis of these mice revealed a loss of GlyR clusters13 and a subset of GABAAR clusters14. Similarly, gene knockouts of single GABAAR subunits, such as the γ2 and α3 subunit, not only interfere with GABAAR but also with gephyrin clustering15,16,17. An X-ray crystal structure of GephE in complex with a 49-residue peptide derived from the large cytoplasmic loop of the GlyR β subunit defined the gephyrin–GlyR interaction in atomic detail10. Recently, we demonstrated that the same region of gephyrin can act as a universal receptor-binding site18, which also mediates the interactions with the GABAAR α1, α2 and α3 subunits19,20,21, and that this interaction is modulated by the oligomeric state of gephyrin together with the number of receptor-binding subunits22, possibly explaining the extremely slow exchange rates of GlyRs at synaptic sites23. Recent studies have highlighted the critical role of the GABAAR α subunits in gephyrin-mediated synaptic versus extrasynaptic targeting of GABAARs, which has been verified in cell-based, electrophysiological24 and immunohistochemical experiments14,19,20,21.
Despite its fundamental function in synaptic receptor localization (Fig. 1), receptor diffusion dynamics20 and synaptic plasticity25, the gephyrin–GABAAR interaction has not yet been elucidated on the molecular level. In this study, we describe the crystal structure of a gephyrin–GABAAR complex and identify key residues in gephyrin and the GABAARs that drive the gephyrin-mediated recruitment of GABAARs to postsynaptic sites. This will allow for a rational approach to develop compounds that could perturb the gephyrin–GABAAR interaction, which will be of great mechanistic and pharmacological interest and could pave the way for new approaches in targeting GABAARs in drug development.
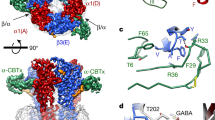
Ionotropic γ-aminobutyric acid type A receptors (GABAARs) are pentameric membrane receptors that are clustered at synaptic sites by direct interactions with the anchoring protein gephyrin. Gephyrin is believed to form higher oligomers at synaptic sites by homotrimerization of its G domain and homodimerization of its E domain. Eleven residues, which are conserved among the synaptic GABAAR α subunits, directly engage with the E domain (PDB entry 4PD0) near the dimer interface (boxes). Gephyrin binding tethers the receptors to the cytoskeleton by direct interaction with profilin and Mena/VASP.
Results
Identification of GABAAR α3-derived minimal peptides
Previous attempts to structurally characterize the interactions between GephE and the full-length intracellular loops of the GABAAR α1, α2 and α3 subunit were hampered by the low affinities and low solubility of the isolated loop regions. Accordingly, a primary challenge was to identify GABAAR-derived peptides appropriate for crystallization. The α3 subunit was selected as a template as it displays the highest in vitro gephyrin affinity of all GABAAR subunits with a KD of 5.3 μM for the full-length intracellular loop18,20,21,26. GABAARs containing α3 subunits co-localize with gephyrin in the cerebellar cortex27, thalamic reticular nucleus17 and at perisomatic synapses in the globus pallidus28, and a knock-out of the α3 subunit results in disruption of postsynaptic gephyrin clusters16,17. In addition, structural insights into the gephyrin-α3 interaction would allow one to predict the interactions with the α1 and α2 subunits to gephyrin, as the binding motifs of these α subunits are conserved18.
Based on the conserved N-terminal gephyrin-binding motif (FS/NIVG) in the GlyR β and GABAAR α3 subunits18, respectively, we synthesized a 20-residue GABAAR α3 fragment (referred to as α20, 367TFNIVGTTYPINLAKDTEFS386). However, the affinity of this 20-mer peptide was too low to be assessed by isothermal titration calorimetry (ITC)18. Hence, we first used peptide SPOT synthesis to perform a complete Ala-scan of the α20 peptide to characterize the molecular details of the GephE–GABAAR α3 interaction. For this assay, GephE was recombinantly expressed, purified and conjugated to horseradish peroxidase (HRP) to allow tracking of its peptide binding by chemiluminescence with high sensitivity. Fourteen different alanine mutants of the α20 peptide were immobilized in high density on a cellulose-based microarray (Celluspot, Intavis). After incubation with GephE-HRP, the chemiluminescence was quantified for each alanine mutant. We observed that Ala substitutions of residues 368–372 as well as 375 significantly impaired binding to GephE, whereas Ala substitutions of Thr373 and, in particular, Thr367 resulted in an increased binding affinity (Fig. 2a).
(a) Peptide-array-based alanine-scan of the gephyrin core-binding site within the GABAAR α3 subunit. Residues shown in pink are conserved in the GABAAR α3 and GlyR β subunits. Gephyrin binding to peptides was detected by chemiluminescence of conjugated horseradish peroxidase. Shown are the relative averaged intensities of six peptide sets together with their standard deviations (error bars). Notably, T367A and T374A increase the gephyrin affinity, whereas an alanine exchange of residues 368–373 as well as residue 376 reduces gephyrin binding. (b) ITC analysis of gephyrin binding to GABAAR α3 and GlyR β/GABAAR α3 chimeric peptides. Peptide sequences and respective GephE affinities are shown. Peptides containing the seven N-terminal residues of the GlyR β peptide display a potentiated gephyrin affinity. N-terminal elongation reduces the affinity, whereas C-terminal elongation has an affinity-enhancing effect. (c) Short GABAAR-derived peptides were sufficient to retain native gephyrin. Pull-down of native gephyrin from whole mouse brain lysate using immobilized short GABAAR-derived peptides. Immunedetection of gephyrin reveals that peptides with a length of 9–11 residues (α9-α11) were sufficient for gephyrin binding, whereas an octamer (α8) did not display binding. The complete blot is shown in Supplementary Fig. 1.
In an attempt to increase the affinity of the GABAAR-derived binding peptide and to map differences in the way GABAAR and GlyR interact with gephyrin, we designed and prepared GABAAR α3/GlyR β chimeric peptides and quantified their binding affinity by ITC (Fig. 2b). We found that the affinity of the α20 peptide could be increased by substituting seven N-terminal residues with those derived from the GlyR β subunit (Fig. 2b, sequences no. 4 and 5), whereas substitution with 13 C-terminal residues from GlyR β did not lead to detectable changes in affinity (Fig. 2b, sequence no. 3).
Based on the Ala-scan and chimeric peptides, we then synthesized optimized variants of GABAAR α3-derived peptides corresponding to the N-terminal region of the GlyR β gephyrin-binding site lacking the N-terminal Thr367 (Fig. 2b, sequences no. 5–10). Strikingly, three GABAAR α3 subunit peptide fragments varying in length from 9 to 11 residues (referred to as α9–11) displayed a micromolar affinity to GephE in ITC experiments (Fig. 2b, sequences no. 7–9). This correlates with the Ala-scan showing that the Thr367Ala mutation increased affinity to GephE (Fig. 2b), and this possibly explains why gephyrin binding of longer GABAAR-derived synthetic peptides could not be quantified by ITC earlier18. Thus, we identified short and soluble GABAAR α3-derived peptides, α9–11, exhibiting micromolar affinity to GephE, which was essential for our overall aim of delineating the gephyrin–GABAAR interaction.
Short GABAAR α3-derived peptides bind native gephyrin
Phosphorylation29,30,31,32, palmitoylation33,34 and acetylation35,36 of gephyrin have been reported and these post-translational modifications have been shown to affect the structure, trafficking, half-life and importantly the ability of gephyrin to interact with partner proteins. To test whether the identified short GABAAR-derived peptide fragments α9–11 also mediate binding to native full-length gephyrin comprising post-translational modifications, the peptides were covalently immobilized on iodoacetyl-activated beads and subsequently incubated with mouse brain lysates. In accordance with the recombinant GephE-based ITC results, α9–11 peptides, but not α8, could retain wild-type gephyrin on the beads, which was verified using the phosphospecific antibody mab7a37 (Fig. 2c and Supplementary Fig. 1). Thus, the identified peptides not only bind to GephE in vitro, but also to full-length wild-type gephyrin from adult mouse brain.
Crystal structures of GephE in complex with GABAAR α3 fragments
Co-crystallization trials with GephE in combination with either the medium-affinity α11WT peptide (FNIVGTTYPIN) or the high-affinity α11SL peptide (FNIVGSLYPIN) yielded four co-crystal structures belonging to two crystal forms (Table 1): (i) hexagonal crystals (space group P61), which were previously described for the GephE-GlyR complex38, diffracting to resolutions of 3.6 and 4.1 Å (Supplementary Fig. 2 and Table 1). (ii) Orthorhombic crystals (space group P21212) diffracting to 2.7 Å resolution (Figs 3 and 4, Supplementary Fig. 3 and Table 1), which were related to GephE-GlyR and apo-GephE structures10,22. The α11WT and α11SL peptides were clearly defined in the electron density maps (Fig. 3c and Supplementary Figs 4 and 5) in both crystal forms, allowing us to model either all or the first nine residues of the 11-mer GABAAR peptide fragment (Figs 3 and 4 and Supplementary Fig. 2).
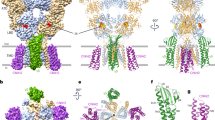
(a) Cartoon representation of GephE in complex with the GABAAR α3-derived peptide α11WT (PDB-ID: 4TK1) colour-coded according to its subdomain architecture as indicated (scheme at the bottom). The residues of α11WT resolved in the structure (368FNIVGTTYP376) are shown as a stick model in orange. (b) Close-up view into the binding pocket. Surface representation of the GephE-binding pocket coloured according to a. The GABAAR peptide is tightly packed into the cleft formed by subdomains III and IV from one monomer, as well as subdomain IV′ from the other monomer. (c) Fo−Fc omit electron density map of the GABAAR α3 peptide (stereo representation) contoured at an rms deviation of 2.5 in blue with the modelled peptide in stick representation.
Close up view of the interactions between GephE with (a) GABAAR α3 wild-type-derived peptide (PDB-ID 4TK1), (b) α11SL peptide (PDB-ID 4TK3) and (c,d) GlyR β-derived peptides (PDB-ID 2FTS10 and 4PD1 (ref. 22)) as well as (e) a superposition and (f,g) schematic 2D representations of the GlyR β wild-type and GABAAR α3 wild-type interactions. In a–d, residues mediating the interactions are highlighted in stick representation and are numbered (coloured for the peptides, black for GephE). GephE residues located in subdomain III are coloured in yellow, residues from subdomain IV in marine blue and GephE residues derived from the other subdomain IV′ in cyan. Hydrogen bonds are shown as dotted red lines. Note that the N-terminal-binding motifs engage in conserved interactions, whereas the C-terminal halves interact differentially with GephE.
The GABAAR α3 and GlyR β-binding sites overlap significantly
The crystal structures of GephE in complex with the GABAAR α3-derived peptide fragments revealed (Fig. 3a) that GephE is present as a homodimer with each subunit consisting of four subdomains as described previously10,22,38. The GephE dimer is in complex with two symmetrically arranged receptor fragments (Fig. 3a), where subdomains III and IV of one GephE monomer as well as subdomain IV of a second monomer (IV′) together from a common receptor-binding site (Figs 3 and 4) for peptides derived from either the GABAAR α3 or the GlyR β subunit10,22,38. The interactions of the peptides derived from either receptor with subdomain III are nearly identical, whereas the interactions with subdomain IV and IV′ are receptor specific (Fig. 4 and Supplementary Fig. 2).
GABAAR α3 and GlyR β also engage in receptor-specific interactions
A direct comparison of the two GephE-α11WT structures with the two GephE-α11SL structures (Fig. 4a,b and Supplementary Fig. 2a,b) as well as the corresponding GlyR β structures (Fig. 4c,d)10,22 revealed distinct interactions with either receptor (Fig. 4e–g). Based on the largely identical N-terminal interactions for α11WT- and GlyR β-derived peptides, we defined the critical N-terminal aromatic residue, which is conserved in all gephyrin-binding receptor-derived peptides as position 1 (Phe368 in GABAAR α3, Phe398 in GlyR β) and based on previous mutational studies18 this residue corresponds to Tyr340 in GABAAR α1 and Tyr339 in GABAAR α2.
The main chains of the conserved N-terminal residues (positions 1–7: GABAAR α3 368FNIVGTT374, GlyR β 398FSIVGSL404 (refs 10)) adopted highly similar conformations (Fig. 4e), whereas their side chains formed subunit-specific interactions with gephyrin (Fig. 4e). The central four-stranded β-sheet of GephE subdomain IV and the first α-helix and several neighbouring residues of subdomain III engaged in identical interactions with the peptides from either the GABAAR α3 or GlyR β subunits (positions 1–5 corresponding to GABAAR 368FNIVG372 and GlyR 398FSIVG402), with the exception of position 2 (Asn369 in GABAAR, Ser399 in GlyR). Notably, the hydrogen-bonded interactions of GABAAR Asn369 with GephE Asp327 appeared to be less optimal when compared with the corresponding hydrogen bonds of Ser399 in GlyR β, thereby contributing to the lower affinity of the GABAAR.
The subsequent residues of the GABAAR and GlyR loops (positions 6–7, GABAAR 373TT374, GlyR 403SL404) interacted with gephyrin differently: GlyR β Ser403 forms additional hydrogen bonds compared with GABAAR Thr373 and, in addition, GABAAR Thr374 could not engage as efficiently with the large hydrophobic pocket formed by Thr716, Tyr719 and Leu722 of gephyrin compared with the corresponding Leu404 in the GlyR β subunit.
The C-terminal residues (positions 8–11: GABAAR 375YPIN378, GlyR 405PRDFEL410) differed substantially in both their side chain interactions and main chain conformations. Nonetheless, GABAAR α3 Tyr375 at position 8 mediates contacts to roughly the same hydrophobic interface on the second GephE monomer as GlyR β Phe408 at position 11. Remarkably, the elongated 49 residue GlyR β loop (β49) forms a short α-helical element, whereas the GABAAR-derived peptides displayed no secondary structure, and, as a result, the overall GephE–GlyR β-binding interface is significant larger (991 Å2) than the corresponding GephE–GABAAR α3-binding interface (835 Å2; Supplementary Tables 1 and 2). This may additionally contribute to the reduced affinity of GABAAR α3 to gephyrin compared with GlyR β.
Thermodynamic dissection of the receptor specificity of gephyrin
Based on the gephyrin-GABAAR α3 X-ray crystal structures, we designed and synthesized a range of point mutated GABAAR α3 peptide fragments and determined the thermodynamic basis of their gephyrin binding by ITC (Fig. 5). Between the structurally resolved GABAAR (368FNIVGTTYPIN378) and GlyR (398FSIVGSLPRDFEL410) fragments, only the four underlined residues are conserved (Fig. 5a). Three of seven non-conserved residues are located at the C-terminal end (GABAAR 376PIN378, GlyR 406RDF408), which is the region where the primary structural differences are found. Nonetheless, exchange of these residues between peptides derived from either GABAAR or GlyR did not alter the overall binding affinity significantly (Fig. 5a), thus indicating comparable binding contributions. Among the four remaining receptor subunit-specific residues, position 2 (GABAAR α3 Asn369, GlyR β Ser399) is located within the conserved N-terminal motif, which mediates the mutually exclusive receptor binding to gephyrin. ITC measurements verified the structural evidence (Fig. 4a–d); Ser399 could engage in more optimal hydrogen-bonding interactions with GephE compared with Asn369. The exchange of Ser399 with an Asn369 in GlyR resulted in an at least threefold reduced affinity (8.3±0.1 to 25.5 μM), vice versa, introduction of Ser for Asn369 in GABAAR increased the affinity at least sixfold (190±30 to 33±6 μM; Fig. 5a). The three remaining subunit-specific residues were located near the centre of the receptor peptide fragments (positions 6–8, GABAAR 373TTY375, GlyR 403SLP405). The ITC analysis revealed that both, the Ser403Thr and the Pro405Tyr exchange, enhanced the peptide fragment affinity (8.3±0.1 to 5.9±0.1 and 8.3±0.1 to 4.3±0.3 μM, respectively), and that this effect was based on additional hydrophobic interactions as reflected by the larger contribution of the entropy term to the overall affinity (Fig. 5a). Exchange of GlyR Leu404 with the corresponding GABAAR Thr reduced the GlyR affinity twofold (8.3±0.1 to 15.9±0.7 μM). In contrast, the corresponding exchange yielded a fivefold higher affinity (190±30 to 36±10 μM) for the GABAAR accompanied by a gain in entropy. This finding was in line with the additional hydrophobic interactions as indicated by the structural analysis (Fig. 4a–d). Remarkably, at position 8, the GlyR and the GABAAR subunit engaged in critical, yet different hydrophobic interactions, with either a Pro in case of the GlyR β subunit or a Tyr in the GABAAR α3 subunit. The critical nature of this ligand position was demonstrated by the corresponding Pro-to-Ala variant, which showed an at least sevenfold reduced affinity (8.3±0.1 to 62±24 μM; Fig. 5a), in line with a loss of a large hydrophobic contact area as indicated by the crystal structure. Finally, a double mutation at positions 2 and 7 within both receptor peptide fragments reduced the GlyR fragment affinity 6-fold (8.3±0.1 to 47±19) and improved the GABAAR fragment affinity 25-fold (190±30 to 7.9±1.3 μM), yielding in the latter case an affinity identical to the GlyR fragment (8.3±0.1 μM; Supplementary Fig. 6). Thus, the ITC experiments provide a detailed picture of the thermodynamic basis of the subunit-specific gephyrin interactions. Together with the X-ray crystal structures, the thermodynamic data complete the picture of how GABAARs and GlyRs are recruited alternatively to postsynaptic sites by engaging with gephyrin as summarized in Fig. 5b.
(a) Different contributions of the binding enthalpy and entropy to the overall free energy. Residues conserved among the GlyR β and GABAAR α3 subunits are shown in magenta, GlyR β residues in blue and GABAAR α3 residues in black. The GlyR β peptide displayed a 23-fold higher affinity than the corresponding GABAAR peptide. Subunit specific residues were exchanged between both receptor subunits. Note that the gephyrin affinity can be maximized by combining hydrophobic residues of GlyRs and GABAARs (GlyR S403T and GABAAR P405Y). Remarkably, full GlyR-like binding affinity can be reconstituted for the GABAAR fragment by only two mutations, N369S and T374L. Enthalpy, entropy and free energy bar graphs are averages (with their standard deviations indicated by the error bars) of at least three ITC measurements. (b) Differential binding of GABAARs and GlyRs to an overlapping gephyrin-binding site. Structures of the GlyR and GABAAR peptides in stick representation colour-coded according to the relative binding contribution (high (>1 kcal mol−1) in red, low (<1 kcal mol−1) in orange and none (~0 kcal mol−1) in blue) with the gephyrin surface shown in grey. The corresponding sequences of the full-length intracellular loops of GABAAR α3 and GlyR β are shown below using the same colour code. Note that different ligand residue positions mediate the critical gephyrin interactions.
Discussion
Despite its fundamental importance for the function of the inhibitory synapse, the interactions with gephyrin that mediate the anchoring and accumulation of synaptic GABAARs were poorly understood. To address this, we designed GABAAR α3-derived gephyrin-binding peptides, which enabled the structural analysis of the gephyrin–GABAAR complex and revealed how GABAARs containing the α3 subunit are clustered at synaptic sites. Despite their binding to an overlapping site in the gephyrin protein compared with GlyRs, GABAARs engage in subunit-specific interactions allowing for an alternative recruitment of inhibitory receptors by gephyrin. In addition, our study identifies two residues (GlyR β Ser399 and Leu404 vs. GABAAR α3 Asn369 and Thr374) as major determinants for gephyrin’s distinct preference for the GlyR β subunit over the GABAAR α3 subunit. Thus, we have provided the structural basis for the mutually exclusive binding of GABAAR and GlyR to the scaffold protein gephyrin18.
Here we present the structural and thermodynamic analysis of gephyrin binding to short peptides, either native to the GABAAR α3 subunit or rationally designed. Notably, the full-length intracellular loop of GABAAR α3 displays an enhanced affinity (KD=5.3 μM)18,21 compared with the truncated α11WT variant (KD=190 μM) analysed here. The 11mer mediates ~70% of the free energy and ~86% of the enthalpy compared with the GABAAR α3 full-length intracellular loop21 when interacting with gephyrin. Although the molecular basis of the enhanced binding cannot be fully explained, the beneficial entropic contribution of the full-length fragment suggests that secondary and/or tertiary structure effects in the full-length intracellular loop present the core-binding site in a more favourable orientation and are thus responsible for its stronger binding. Furthermore, the α11WT peptide (368FNIVGTTYPIN378) occupies an overlapping binding site (Fig. 4e) compared with the GlyR fragment resolved earlier (398FSIVGSLPRDFELS411), which, in turn, could be used to completely block gephyrin binding of GABAAR-derived full-length intracellular loops18. In addition, single point-mutations within the core region of the α3-derived peptide (368FNIVGTTY375) were shown18 to completely abolish gephyrin binding of the full-length loop and, vice versa, loops missing the core motif were demonstrated to completely lose their ability to mediate receptor clustering in neurons or to mediate binding in yeast two-hybrid experiments21. Taken together, it can be concluded that the remainder of the intracellular loop exerts a critical enhancing effect, but does not provide a second independent binding site that would be sufficient for binding in primary hippocampal neurons, yeast two-hybrid, ITC or pull-down assays.
A direct comparison of the gephyrin affinity to the core-binding sites of GlyR and GABAAR confirmed an approximately 25-fold preference of gephyrin for GlyRs containing the β subunit over GABAARs containing the α3 subunit. The structural and thermodynamic mutational analysis revealed that Ser399 of the GlyR binds more effectively than the corresponding Asn369 in the GABAAR α3 subunit, whereas Leu404 of the GlyR creates a larger hydrophobic binding interface than the corresponding GABAAR α3 residue Thr374 (Fig. 4 and Supplementary Fig. 6). Together, the slight differences in the side chain interactions synergistically result in the distinct subunit preference of gephyrin for GlyRs over α3-containing GABAARs.
Gephyrin mediates GABAAR α3 and GlyR β binding via a large groove formed by subdomains III and IV (Fig. 3b) within GephE. The N-terminal part of the receptor core-binding sites relies on highly conserved receptor interactions and involves hydrophobic contacts between Phe330 of gephyrin and the respective aromatic residues in the receptors, but also critical hydrogen bonds mediated by Tyr673 and Asp327 of gephyrin. We observed that the seven N-terminal residues contribute the majority of the overall receptor-binding strength, and that the peptide-binding motif derived from the GlyR β subunit displays the highest gephyrin-binding potency. GlyR and GABAAR co-localization and agonist co-transmission were reported earlier39,40. It was shown that up to 35% of all GlyRs co-localize with GABAARs in the hypoglossal nucleus39 and that 20–40% of all miniature postsynaptic currents recorded from respiratory glycinergic neurons are mixed miniature postsynaptic currents40 that result from the co-release of GABA and glycine. The competition between GABAARs and GlyRs for an overlapping gephyrin-binding site, as shown here, suggests an interdependence of their clustering as well as their transport to the synapse. Major determinants of the competition would be the ratio of free receptor-binding sites in gephyrin and, among the gephyrin-binding receptor subunits, the gephyrin-binding subunit number within a pentameric receptor and the post-translational modifications of the respective motifs within these subunits20,41. Although GABAAR transport remains poorly characterized, GlyRs were shown to be retrogradely co-transported with gephyrin by the dynein motor complex via an interaction of the dynein light chain with the central linker of gephyrin42. If GABAARs would rely on the same pathway for their retrograde transport, both inhibitory receptor families would also compete for a common transport pathway, thus further intertwining their presence at inhibitory synapses. Finally, a recent study of the ultrastructure of spinal cord inhibitory synapses suggested that the GABAAR/GlyR competition for gephyrin-binding sites is regulated in an activity-dependent manner23.
Overall, the GephE-peptide structures display a high similarity. However, two out of eight peptide chains in the GephE–peptide complexes derived from the orthorhombic and hexagonal space groups exhibit obvious differences in the orientation of GABAAR Tyr375 (Supplementary Fig. 3). We find that these changes are due to crystal contacts with a neighbouring symmetry-related molecule and therefore do not indicate a difference between the two receptor-binding sites within a GephE dimer as suggested earlier38. Accordingly, our structures clearly support the view that both receptor-binding sites within a single GephE dimer are identical and hence are occupied in a non-cooperative manner.
Our structural analysis demonstrates that the C-terminal halves of the GlyR and GABAAR gephyrin-binding core regions interact differently with gephyrin and occupy partially non-identical binding surfaces. The analysis of the chimeric peptide variants demonstrated that the C-terminal regions are crucial for the binding event by exhibiting an important enhancing effect. Furthermore, the structural and thermodynamic dissections revealed that the side chain interactions of GABAAR α3 residues Thr373, Thr374 and Tyr375 within this C-terminal receptor region are of major importance. Particularly, Tyr375, which is conserved among the α1, α2 and α3 GABAAR subunits, engages in critical hydrophobic interactions. Based on the sequence similarity of the gephyrin-binding region of the GABAAR α3 and α1 subunits and a previous mutagenesis study18, the gephyrin-binding interface of the GABAAR α1 subunit can be predicted (Supplementary Fig. 7).
The fact that different receptor subunits target gephyrin with unique binding motifs, which nonetheless engage in tight and specific interactions, allows for a subunit-specific post-translational regulation of this interaction. To this end, our study provides a possible structural explanation for the previously described regulation of GABAergic transmission by the extracellular signal-regulated kinases/mitogen-activated protein kinases pathway mediated by phosphorylation of GABAAR α1 Thr345, which is conserved within the gephyrin-binding site of GABAAR α1–3 and corresponds to GABAAR α3 Thr373 (Supplementary Fig. 7)43. Furthermore, we provide a structural framework for future functional studies of phosphorylations at GABAAR α3 Thr374 and Tyr375 and their corresponding GABAAR α1 and α2 residues, which are phosphorylated in vivo44,45. It can be assumed that post-translational modifications of these residues modulate the gephyrin-GABAAR affinity, and hence, the residence time at the synapse in a similar manner as reported for the protein kinase C-mediated phosphorylation of GlyR β Ser399 (ref. 41).
In contrast to glutamate receptors for which bacterial homologues have been identified, no GlyR-related receptors have been identified in bacteria. Furthermore, GlyRs are also absent from invertebrates46 and, in agreement with this observation, only those residues that are required for the receptor interaction are conserved among gephyrin proteins from vertebrates10. In contrast, GABAAR α subunit-related proteins have been identified in invertebrates46 and, in order to decipher how the gephyrin-binding motif evolved within the GABAAR family, we compared the corresponding sequences (Supplementary Fig. 8). Similar to the gephyrin-binding motif of GlyRs, only vertebrate GABAARs, but not invertebrate GABAARs, contain a largely conserved gephyrin-binding sequence. This suggests that gephyrin-mediated GABAAR clustering evolved later than gephyrin-mediated GlyR clustering and that both motifs evolved independently.
As major mediators of fast synaptic inhibition GABAARs are clinically relevant drug targets, and molecules that uncouple the central receptor-scaffold interaction at post-synaptic sites would therefore provide a powerful pharmacological tool with a possible therapeutic relevance. This principle has been demonstrated for the interaction between the N-methyl-D-aspartate-type glutamate receptors and the scaffolding postsynaptic density protein 95 (PSD-95), which has been targeted by peptide-based inhibitors47,48,49,50,51,52. This approach has shown great promise both as a pharmacological tool51,53 and, in particular, in the development of therapeutically relevant compounds54,55. We therefore propose that conceptually similar molecules could be used to interfere with the receptor-scaffold interactions in vivo to modulate GABAergic and/or glycinergic transmission. Molecules that bind the universal N-terminal receptor-binding site with high affinity could be used as competitive inhibitors of gephyrin-mediated synaptic GlyR and GABAAR clustering. In addition, the described alternative receptor recruitment of GABAARs versus GlyRs by gephyrin, could form the basis for the development of subunit-specific modulators of either GABAergic or glycinergic transmission.
Methods
Peptide synthesis, purification and characterization
Apart from four chimeric peptides, which were purchased as lyophilized powder from Genscript, peptides were synthesized using Fmoc solid phase peptide synthesis. Preparative HPLC was performed on an Agilent 1100 system using a C18 reverse phase column (Zorbax 300 SB-C18, 21.2 × 250 mm2) with a linear gradient of the binary solvent system of 0.1% trifluoroacetic acid in H2O/acetonitrile (ACN; A: 95:5 and B: 5:95) with a flow rate of 20 ml min−1. Analytical HPLC was performed on an Agilent 1100 system with a C18 reverse phase column (Zorbax 300 SB-C18 column, 4.6 × 150 mm2), a flow rate of 1 ml min−1, and a linear gradient of the binary solvent system of 0.1% trifluoroacetic acid in H2O/ACN (A: 95:5 and B: 5:95). Mass spectra were obtained with an Agilent 6410 Triple Quadrupole Mass Spectrometer instrument using electron spray ionization coupled to an Agilent 1200 HPLC system (electrospray ionization–liquid chromatography/ mass spectrometry (ESI-LC/MS)) with a C18 reverse phase column (Zorbax Eclipse XBD-C18, 4.6 × 50 mm), autosampler and diode-array detector using a linear gradient of the binary solvent system of 0.1% formic acid in H2O/ACN (A: 95:5 and B: 5:95) with a flow rate of 1 ml min−1. During ESI-LC/MS analysis, evaporative light scattering traces were obtained with a SedereSedex 85 Light Scattering Detector. The identity of all tested compounds was confirmed by ESI-LC/MS (Supplementary Table 3), which also provided purity data (all >90%; ultraviolet and evaporative light scattering detection). All used peptides were water soluble at neutral pH in the millimolar range.
Protein expression and purification
GephE (gephyrin P1 splice variant residues 318–736) as well as residues 378–425 of the large cytoplasmic loop of the GlyR β subunit (β49) were expressed in E. coli BL21 (DE3) (Stratagene) as an intein fusion proteins with a chitin-binding domain for affinity purification9. Cells were grown in lysogeny broth medium at 30 °C and induced with 0.5–1 mM isopropyl-β-thiogalactoside at a cell density A600 of 0.5–1. Cells were collected after 4 h (Intein-GlyR) or 20 h (GephE-Intein) by centrifugation (4,000 g), resuspended in lysis buffer (50 mM Tris/HCl, pH 8.0, 500 mM NaCl) and passed through a cell disruptor (Constant Systems). Cell debris was removed by centrifugation (70,000 g). Proteins were purified at room temperature (RT) using a chitin column according to the instructions of the IMPACT-TWIN protein expression and purification system (New England Biolabs). Cleavage of GephE from the N-terminal intein fusion was induced by a pH and temperature shift (100 mM NaCl, 20 mM Tris/HCl, 5 mM DTT, 1 mM EDTA, pH 6, 25 °C) and subsequent incubation for 48 h. Cleavage of β49 from the intein was accomplished by incubation with 100 mM DTT for 24 h. Protein containing fractions were collected, concentrated and applied to a 26/60 Superdex 200 size exclusion column (Amersham Biosciences) equilibrated with buffer (10 mM Tris/HCl, pH 8.0, 250 mM NaCl, 1 mM β-mercaptoethanol). Pure fractions were pooled, concentrated using Vivaspin 3 kDa molecular weight cutoff centrifugal filter devices (Sartorius Stedim Biotech) to 1–100 mg ml−1, flash-frozen in 0.5 ml aliquots and stored at −80 °C.
Protein conjugation for chemiluminescence detection
Purified GephE was conjugated with HRP (Sigma-Aldrich) to enable its detection by chemiluminescence. The procedure is based on earlier reports56. In brief, 5 mg of lyophilized HRP were dissolved in 1 ml conjugation buffer (50 mM Na2CO3, pH 8.0). After incubation with 1 ml of 50 mM NaIO4 for 30 min and 1 ml of 160 mM ethylene glycol for 1 h, the reaction mixture was dialysed against 10 mM Na2CO3, pH 9.5 at 4 °C overnight. 5 mg of GephE were added to the dialysed activated HRP containing solution and after incubation for 3 h, 5 mg of NaBH4 were added and the reaction was stirred for another 3 h at 4 °C. After dialysis against SEC-buffer (10 mM Tris/HCl pH 8.0, 250 mM NaCl, 1 mM β-mercaptoethanol), the solution was centrifuged for 5 min at 10,000 g and the supernatant was applied to a 26/60 Superdex 200 size exclusion column (Amersham Biosciences) equilibrated with SEC-buffer. Pure fractions of the GephE-HRP-conjugate were identified via SDS–polyacrylamide gel electrophoresis and pooled according to chemiluminescence and concentrated using Vivaspin 3 kDa molecular weight cutoff centrifugal filter devices (Sartorius Stedim Biotech) to 1 mg ml−1, flash-frozen in 0.1 ml aliquots and stored at −80 °C.
Peptide array-based analysis
The wild-type GABAAR α3 peptide (369TFNIVGTTYPINLAK384) and its respective Ala-variants were purchased from Intavis AG in a CelluSpot format. After rinsing of the peptide array slides with TBS (50 mM Tris, 150 mM NaCl, pH 7.6) and 0.05% Tween-20 (TBST) for 5 min, the peptide array was incubated with HRP-conjugated GephE (1 μM) in blocking buffer (TBST+5% nonfat dry milk) for 5 h at RT. After extensive washing, bound GephE-HRP-conjugate was detected by chemiluminescence using the Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) using the chemiluminescent bio-imaging system MicroChemi (DNR Bio-imaging Systems). The resulting dot-blots were analysed using the array analyse software (Active Motif). Three peptide array duplicates were used to calculate the average spot densities and their rms deviations.
Preparation of whole brain lysates
After cervical dislocation, whole brains from 54-week-old C57Bl/6J male mice were removed from the scull and rapidly homogenized in 1 ml lysate buffer (20 mM HEPES, 100 mM K-acetate, 40 mM KCl, 5 mM EGTA, 5 mM MgCl2, 5 mM DTT, 1 mM PMSF, 1% Triton X, protease inhibitor Roche complete, pH 7.2) per 200 mg using a pistol homogenizer (8 strokes at 900 r.p.m.). The homogenate was centrifuged at 10,000g for 15 min. Subsequently, the supernatant was removed and aliquots were flash-frozen in liquid nitrogen and stored at −80 °C.
Covalent immobilization of peptides
GABAAR α3 peptide fragments with an additional C-terminal Cys (368FNIVGTTY375C, 368FNIVGTTYP376C, 368FNIVGTTYPI377C, 368FNIVGTTYPIN378C) were coupled to UltraLinkIodoacetyl Gel (Thermo Scientific) according to the protocol of the manufacturer: the peptides were dissolved in coupling buffer (50 mM Tris, 5 mM EDTA, pH 8.5) at a concentration of 1 mM and incubated for 2 h at RT with UltraLink beads, which had been washed and equilibrated with coupling buffer before. After removing excess peptides, the UltraLink beads were subjected to 1 mM cysteine for 2 h to quench possible unreacted iodoacetyl groups. The resin was washed three times and equilibrated with 1 M NaCl and stored at 4 °C.
Pull-down and western blot detection
The resin with the immobilized peptides was incubated with brain lysate for 1 h at 4 °C. After three washing steps with lysate buffer, the beads were boiled with Laemmli buffer containing 10% SDS. Subsequently, the supernatant was applied to an SDS–polyacrylamide gel electrophoresis followed by western blotting against gephyrin using the mAb7a antibody37 (Synaptic Systems) at a dilution of 1:500.
Peptide and protein concentration determination
The concentration of the GephE stock-solution was determined by amino-acid analysis and aliquots of an identical stock were used for all experiments to ensure comparability of all experiments and rule out effects of protein activity, degradation, concentration determination and aggregation. Peptide stocks were prepared by weighing the lyophilized powders. All ITC titrations displayed stoichiometries between 0.96 and 1.04, thus, demonstrating a high accuracy of the measured concentrations and comparability of the results.
Isothermal titration calorimetry
The experiments were performed using an ITC200 (MicroCal) at 25 °C and 1,000 r.p.m. stirring and designed so that c-values were generally within 0.5–100 (c-value=KA × [protein] × N with KA, equilibrium association constant; [protein], protein concentration; N, stoichiometry). Specifically, 40 μl of a solution containing 1–3 mM of the peptide were titrated into the 200 μl sample cell containing 25–100 μM GephE. In each experiment, a volume of 1–2 μl of ligand was added at a time resulting in 20–40 injections and a final molar ratio between 1:3 and 1:6. Ligand-to-buffer titrations were carried out in an analogous manner, so that the heat produced by injection, mixing and dilution could be subtracted. The binding enthalpy was directly measured, whereas the dissociation constant (KD) and stoichiometry (N) were obtained by data analysis using the Origin software (OriginLab). Measurements were conducted at least three times and are given as mean values with the resulting standard deviations.
Protein complex crystallization and X-ray data collection
Complexes of GephE and peptides α11WT and α11SL were prepared by mixing both in a 1:5 (protein/peptide) molar ratio followed by incubation at 4 °C for 15 min. The complexes were crystallized by the hanging drop vapour diffusion method at 20 °C at a concentration of 10 mg ml−1 in the presence of 0.2 M calcium acetate, 0.1 M MES, pH 6, and 6–10% isopropanol as precipitant. Both complexes were also crystallized at 4 °C at a concentration of 2.5 mg ml−1 with 0.1 M Tris, pH 7.5, and 21–27% PEG4000 as precipitant. The crystals were transferred to the respective mother liquor solution containing 25% glycerol as cryoprotectant and flash frozen in liquid nitrogen. The data sets were collected at beam lines ID14–4 and ID 23-2 at the ESRF, Grenoble (Table 1).
Structure determination and refinement
The data sets were indexed and integrated with iMosflm57 or XDS58, further scaling and merging were done by using the CCP4 suite59. The structures were solved by molecular replacement with Phaser60 using 2FU3 as initial model10. Refinements were carried out with PHENIX61 and Refmac5 (ref. 59). The crystals grown at 4 °C belonged to space group P21212 and diffracted to a resolution of 2.7 Å, the crystals grown at 20 °C to space group P61, which diffracted only to low resolutions of 3.6–4.1 Å. For low-resolution refinement, the 1.7-Å resolution apo-E domain structure (PDB: 4PDO) was used as reference model. Figures involving molecular representations were prepared using PyMol ( http://www.pymol.org).
Additional information
How to cite this article: Maric, H. M. et al. Molecular basis of the alternative recruitment of GABAA versus glycine receptors through gephyrin. Nat. Commun. 5:5767 doi: 10.1038/ncomms6767 (2014).
Accession codes: The coordinates have been deposited in the Protein Data Bank with the following codes: 4TK1 (orthorhombic) and 4TK2 (hexagonal) for GephE-α11WT, 4TK3 (orthorhombic) and 4TK4 (hexagonal) for GephE-α11SL structures.
References
Hines, R. M., Davies, P. A., Moss, S. J. & Maguire, J. Functional regulation of GABAA receptors in nervous system pathologies. Curr. Opin. Neurobiol. 22, 552–558 (2012).
Kittler, J. T., McAinsh, K. & Moss, S. J. Mechanisms of GABAA receptor assembly and trafficking: implications for the modulation of inhibitory neurotransmission. Mol. Neurobiol. 26, 251–268 (2002).
Tretter, V. et al. Gephyrin, the enigmatic organizer at GABAergic synapses. Front. Cell Neurosci 6, 23 (2012).
Tyagarajan, S. K. & Fritschy, J. M. Gephyrin: a master regulator of neuronal function? Nat. Rev. Neurosci. 15, 141–156 (2014).
Fritschy, J. M. & Panzanelli, P. GABA receptors and plasticity of inhibitory neurotransmission in the central nervous system. Eur. J. Neurosci. 39, 1845–1865 (2014).
Pfeiffer, F., Graham, D. & Betz, H. Purification by affinity chromatography of the glycine receptor of rat spinal cord. J. Biol. Chem. 257, 9389–9393 (1982).
Meyer, G., Kirsch, J., Betz, H. & Langosch, D. Identification of a gephyrin binding motif on the glycine receptor β subunit. Neuron 15, 563–572 (1995).
Kneussel, M., Hermann, A., Kirsch, J. & Betz, H. Hydrophobic interactions mediate binding of the glycine receptor β-subunit to gephyrin. J. Neurochem. 72, 1323–1326 (1999).
Schrader, N. et al. Biochemical characterization of the high affinity binding between the glycine receptor and gephyrin. J. Biol. Chem. 279, 18733–18741 (2004).
Kim, E. Y. et al. Deciphering the structural framework of glycine receptor anchoring by gephyrin. EMBO J. 25, 1385–1395 (2006).
Kirsch, J. et al. The 93-kDa glycine receptor-associated protein binds to tubulin. J. Biol. Chem. 266, 22242–22245 (1991).
Giesemann, T. et al. Complex formation between the postsynaptic scaffolding protein gephyrin, profilin, and Mena: a possible link to the microfilament system. J. Neurosci. 23, 8330–8339 (2003).
Feng, G. et al. Dual requirement for gephyrin in glycine receptor clustering and molybdoenzyme activity. Science 282, 1321–1324 (1998).
Kneussel, M. et al. Loss of postsynaptic GABAA receptor clustering in gephyrin-deficient mice. J. Neurosci. 19, 9289–9297 (1999).
Essrich, C., Lorez, M., Benson, J. A., Fritschy, J. M. & Luscher, B. Postsynaptic clustering of major GABAA receptor subtypes requires the γ2 subunit and gephyrin. Nat. Neurosci. 1, 563–571 (1998).
Studer, R. et al. Alteration of GABAergic synapses and gephyrin clusters in the thalamic reticular nucleus of GABAA receptor α3 subunit-null mice. Eur. J. Neurosci. 24, 1307–1315 (2006).
Winsky-Sommerer, R. et al. Normal sleep homeostasis and lack of epilepsy phenotype in GABAA receptor α3 subunit-knockout mice. Neuroscience 154, 595–605 (2008).
Maric, H. M., Mukherjee, J., Tretter, V., Moss, S. J. & Schindelin, H. Gephyrin-mediated γ-aminobutyric acid type A and glycine receptor clustering relies on a common binding site. J. Biol. Chem. 286, 42105–42114 (2011).
Tretter, V. et al. The clustering of GABAA receptor subtypes at inhibitory synapses is facilitated via the direct binding of receptor α2 subunits to gephyrin. J. Neurosci. 28, 1356–1365 (2008).
Mukherjee, J. et al. The residence time of GABA(A)Rs at inhibitory synapses is determined by direct binding of the receptor α1 subunit to gephyrin. J. Neurosci. 31, 14677–14687 (2011).
Tretter, V. et al. Molecular basis of the gamma-aminobutyric acid A receptor α3 subunit interaction with the clustering protein gephyrin. J. Biol. Chem. 286, 37702–37711 (2011).
Maric, H. M., Kasaragod, V. B. & Schindelin, H. Modulation of gephyrin-glycine receptor affinity by multivalency. ACS Chem. Biol doi:10.1021/cb500303a (2014).
Specht, C. G. et al. Quantitative nanoscopy of inhibitory synapses: counting gephyrin molecules and receptor binding sites. Neuron 79, 308–321 (2013).
Wu, X. et al. γ-Aminobutyric acid type A (GABAA) receptor α subunits play a direct role in synaptic versus extrasynaptic targeting. J. Biol. Chem. 287, 27417–27430 (2012).
Vlachos, A., Reddy-Alla, S., Papadopoulos, T., Deller, T. & Betz, H. Homeostatic regulation of gephyrin scaffolds and synaptic strength at mature hippocampal GABAergic postsynapses. Cereb. Cortex 23, 2700–2711 (2013).
Kowalczyk, S. et al. Direct binding of GABAA receptor β2 and β3 subunits to gephyrin. Eur. J. Neurosci. 37, 544–554 (2013).
Sassoe-Pognetto, M., Panzanelli, P., Sieghart, W. & Fritschy, J. M. Colocalization of multiple GABAA receptor subtypes with gephyrin at postsynaptic sites. J. Comp. Neurol. 420, 481–498 (2000).
Gross, A. et al. Differential localization of GABAA receptor subunits in relation to rat striatopallidal and pallidopallidal synapses. Eur. J. Neurosci. 33, 868–878 (2011).
Tyagarajan, S. K. et al. Extracellular signal-regulated kinase and glycogen synthase kinase 3 β regulate gephyrin postsynaptic aggregation and GABAergic synaptic function in a calpain-dependent mechanism. J. Biol. Chem. 288, 9634–9647 (2013).
Herweg, J. & Schwarz, G. Splice-specific glycine receptor binding, folding, and phosphorylation of the scaffolding protein gephyrin. J. Biol. Chem. 287, 12645–12656 (2012).
Demirkan, G., Yu, K., Boylan, J. M., Salomon, A. R. & Gruppuso, P. A. Phosphoproteomic profiling of in vivo signaling in liver by the mammalian target of rapamycin complex 1 (mTORC1). PLoS ONE 6, e21729 (2011).
Zita, M. M. et al. Post-phosphorylation prolyl isomerisation of gephyrin represents a mechanism to modulate glycine receptors function. EMBO J. 26, 1761–1771 (2007).
Kang, R. et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909 (2008).
Dejanovic, B. et al. Palmitoylation of gephyrin controls receptor clustering and plasticity of GABAergic synapses. PLoS Biol. 12, e1001908 (2014).
Schwer, B. et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell 8, 604–606 (2009).
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009).
Kuhse, J. et al. Phosphorylation of gephyrin in hippocampal neurons by cyclin-dependent kinase CDK5 at Ser-270 is dependent on collybistin. J. Biol. Chem. 287, 30952–30966 (2012).
Sola, M. et al. Structural basis of dynamic glycine receptor clustering by gephyrin. EMBO J. 23, 2510–2519 (2004).
Muller, E., Le Corronc, H., Triller, A. & Legendre, P. Developmental dissociation of presynaptic inhibitory neurotransmitter and postsynaptic receptor clustering in the hypoglossal nucleus. Mol. Cell Neurosci. 32, 254–273 (2006).
Rahman, J., Latal, A. T., Besser, S., Hirrlinger, J. & Hulsmann, S. Mixed miniature postsynaptic currents resulting from co-release of glycine and GABA recorded from glycinergic neurons in the neonatal respiratory network. Eur. J. Neurosci. 37, 1229–1241 (2013).
Specht, C. G. et al. Regulation of glycine receptor diffusion properties and gephyrin interactions by protein kinase C. EMBO J. 30, 3842–3853 (2011).
Maas, C. et al. Neuronal cotransport of glycine receptor and the scaffold protein gephyrin. J. Biol. Chem. 172, 441–451 (2006).
Bell-Horner, C. L., Dohi, A., Nguyen, Q., Dillon, G. H. & Singh, M. ERK/MAPK pathway regulates GABAA receptors. J. Neurobiol. 66, 1467–1474 (2006).
Munton, R. P. et al. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol. Cel. Proteomics 6, 283–293 (2007).
Ballif, B. A., Carey, G. R., Sunyaev, S. R. & Gygi, S. P. Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J. Proteome Res. 7, 311–318 (2008).
Xue, H. Identification of major phylogenetic branches of inhibitory ligand-gated channel receptors. J. Mol. Evol. 47, 323–333 (1998).
Bach, A. et al. Modified peptides as potent inhibitors of the postsynaptic density-95/N-methyl-D-aspartate receptor interaction. J. Med. Chem. 51, 6450–6459 (2008).
Bach, A. et al. Cell-permeable and plasma-stable peptidomimetic inhibitors of the postsynaptic density-95/N-methyl-D-aspartate receptor interaction. J. Med. Chem. 54, 1333–1346 (2011).
Bach, A. et al. Design and synthesis of highly potent and plasma-stable dimeric inhibitors of the PSD-95-NMDA receptor interaction. Angew. Chem. Int. Ed. 48, 9685–9689 (2009).
Bach, A. et al. A high-affinity, dimeric inhibitor of PSD-95 bivalently interacts with PDZ1-2 and protects against ischemic brain damage. Proc. Natl Acad. Sci. USA 109, 3317–3322 (2012).
Sainlos, M. et al. Biomimetic divalent ligands for the acute disruption of synaptic AMPAR stabilization. Nat. Chem. Biol. 7, 81–91 (2011).
Iskenderian-Epps, W. S. & Imperiali, B. Modulation of Shank3 PDZ domain ligand-binding affinity by dimerization. Chembiochem 11, 1979–1984 (2010).
Chi, C. N., Bach, A., Strømgaard, K., Gianni, S. & Jemth, P. Ligand binding by PDZ domains. BioFactors 38, 338–348 (2012).
Flemming, A. Stroke: Can PSD95 inhibitors widen the therapeutic window? Nat. Rev. Drug Discov. 11, 272 (2012).
Dolgin, E. To serve and neuroprotect. Nature Med 18, 1003–1006 (2012).
Nakane, P. K. & Kawaoi, A. Peroxidase-labeled antibody. A new method of conjugation. J Histochem. Cytochem. 22, 1084–1091 (1974).
Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Bailey, S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (H.S.: FZ 82, SFB 487 C7, SCHI 425/8-1 and M.K.: GRK1459 and KN 556/6-1), H.M.M. was supported by Lundbeckfonden (R118-A11469) and a grant of the German Excellence Initiative to the Graduate School of Life Sciences, University of Würzburg.
Author information
Authors and Affiliations
Contributions
H.M.M. carried out all experiments except the crystallization and structure determination, which was carried out by V.B.K., and the preparation of brain lysates, pull-down and WB experiments, which were carried out by T.J.H. Research was designed by H.M.M., T.J.H., H.S., K.S., M.K. and V.T. The manuscript was written by H.M.M., V.B.K. and H.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-8, Supplementary Tables 1-3, Supplementary Discussion and Supplementary References. (PDF 1423 kb)
Rights and permissions
About this article
Cite this article
Maric, H., Kasaragod, V., Hausrat, T. et al. Molecular basis of the alternative recruitment of GABAA versus glycine receptors through gephyrin. Nat Commun 5, 5767 (2014). https://doi.org/10.1038/ncomms6767
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms6767
This article is cited by
-
Spectrin-beta 2 facilitates the selective accumulation of GABAA receptors at somatodendritic synapses
Communications Biology (2023)
-
Complex regulation of Gephyrin splicing is a determinant of inhibitory postsynaptic diversity
Nature Communications (2022)
-
Human ARHGEF9 intellectual disability syndrome is phenocopied by a mutation that disrupts collybistin binding to the GABAA receptor α2 subunit
Molecular Psychiatry (2022)
-
Multivalent binding kinetics resolved by fluorescence proximity sensing
Communications Biology (2022)
-
Mesophasic organization of GABAA receptors in hippocampal inhibitory synapses
Nature Neuroscience (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.