Abstract
A key feature of mitochondrial translation is the reduced number of transfer RNAs and reassignment of codons. For human mitochondria, a major unresolved problem is how the set of stop codons are decoded by the release factors mtRF1a and mtRF1. Here we present three-dimensional structural models of human mtRF1a and mtRF1 based on their homology to bacterial RF1 in the codon recognition domain, and the strong conservation between mitochondrial and bacterial ribosomal RNA in the decoding region. Sequence changes in the less homologous mtRF1 appear to be correlated with specific features of the mitochondrial rRNA. Extensive computer simulations of the complexes with the ribosomal decoding site show that both mitochondrial factors have similar specificities and that neither reads the putative vertebrate stop codons AGA and AGG. Instead, we present a structural model for a mechanism by which the ICT1 protein causes termination by sensing the presence of these codons in the A-site of stalled ribosomes.
Similar content being viewed by others
Introduction
The mitochondrion is an organelle found in most eukaryotic cells with critical functions for ATP production. Although the mitochondrion has its own DNA and translation system1, the human mitochondrial DNA contains only 37 genes encoding the two ribosomal RNAs, 22 transfer RNAs and 13 polypeptides2. All other proteins, including translation factors and ribosomal proteins, are imported from the cytosol. The mitoribosome is in many ways similar to bacterial ribosomes and several of its translation factors show activity on these3. This in line with the fact that the key functional regions involved in decoding and peptide bond formation are largely conserved4, which they are most likely to be also on the tertiary structure level. Nevertheless, the overall composition and structure of the mitoribosome differs considerably from its bacterial counterpart, for example, with roughly inverted protein-to-RNA ratio5 of about 3:1 and a larger total mass6.
Although evolution has led to a universal genetic code, many examples of deviations exist7,8. In vertebrate mitochondria, the standard stop codon UGA has been reassigned to encode tryptophan, whereas the two arginine codons AGA and AGG have no cognate tRNAs and have instead been reassigned to stop codons in many organisms2. For example, in human mitochondria the CO1 and ND6 genes end with AGA and AGG stop codons, respectively2. Interestingly, four different human proteins homologous to the bacterial release factor RF1 have been found9: mtRF1, mtRF1a, ICT1 and C12orf65. mtRF1 was identified by bioinformatics and was initially believed to read all four of the mitochondrial stop codons10. A new candidate, mtRF1a, was later found and shown to have release activity in vitro on Escherichia coli ribosomes for the UAA and UAG stop codons11,12. However, no release activity for mtRF1 was observed at any of the mitochondrial stop codons UAA, UAG, AGA and AGG in these bacterial termination assays. Yet, the possibility of mtRF1 being responsible for reading the non-standard stop codons on the mitoribosome cannot be excluded13, as it has two insertions in the region reading the first stop codon position14, which might confer the ability to read an adenine. These insertions consist of three amino acids (Gly–Leu–Ser) in the stop codon recognition loop and two residues (Arg–Thr) in the short loop preceding the α5 helix. The other two homologues of the bacterial release factors (RFs), ICT1 and C12orf65, are both mitochondrial proteins with critical functions for cell viability and have been proposed to be involved in rescuing stalled ribosomes9,15,16. ICT1 has further been shown to be an integral part of the mitoribosome9. However, both proteins lack the codon recognition domain of RF1 and RF2, which is present in mtRF1 and mtRF1a.
The termination mechanism in mitochondria thus remains an unsolved problem and it is still unclear whether there are one or two mitochondrial release factors (mtRFs) and whether there actually are two or four stop codons in vertebrate mitochondria. Previous studies have suggested two different hypotheses for the termination at non-standard stop codons, where the first invokes RNA editing11 and the second a −1 frameshift17, both leading to the mtRFs effectively reading a UAG codon. Neither of these hypotheses can be completely verified or disproved without an in vitro vertebrate mitochondrial translation assay, but no such system is available at present. However, computational modelling and molecular dynamics (MD) simulations have been remarkably successful in predicting key features of several steps involved in the translation process18, including termination by class-1 RFs19,20, and may offer a useful strategy for exploring the mitochondrial termination problem.
Here we first employ homology modelling based on the structure of RF1 in complex with the bacterial ribosome14 to derive structural models of mtRF1a and mtRF1. These models are then used to determine codon specificities through extensive MD simulations and free-energy calculations. The results show that neither of the two factors is able to read AGA or AGG. Instead, we present an alternative mechanism by which ICT1 causes termination.
Results
Homology modelling
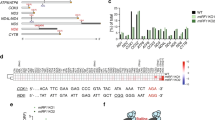
Homology models of mtRF1a and mtRF1 were created with automated protocols using the crystal structure of the Thermus thermophilus RF1–ribosome complex14 as a template, including the stop codon. Sequence alignments of the RF regions close to the stop codon and the ribosomal A-site are shown in Fig. 1 together with the predicted three-dimensional structures. The overall sequence identities for mtRF1a and mtRF1 with RF1 are 47% and 39%, respectively, and the values for the stop codon recognition domain are very similar. mtRF1a has a high sequence identity with bacterial RF1 in the stop codon recognition region (Fig. 1a) and could thus be expected to have analogous codon-reading properties. mtRF1, on the other hand, is similar to RF1 around the third stop codon position but is distinctly different near the first and second positions, where the two insertions are present. As mentioned above, mtRF1 has an RT insertion just before the α5 helix and a GLS insertion in the recognition loop (Fig. 1c). Because of these insertions, it appears essential to include (the conserved) rRNA bases close to the recognition loop in the template to avoid unrealistic loop conformations. It was further noted that A1913 in the bacterial large subunit helix 69, interacting with RF1, corresponds to a C in the human mitoribosome, and this substitution was also included in the template.
(a) Sequence alignments with T. thermophilus RF1 used in the homology modelling of mtRF1a and mtRF1. Residues involved in first position reading are coloured in green (note that these are backbone interactions) and those involved in the second and third position reading are shown in cyan. (b) RF1 crystal structure in the ribosome complex14. (c) Close-up image of the decoding region with the homology models of mtRF1a (cyan) and mtRF1 (magenta) superimposed on the crystal structure of the RF1–ribosome complex (yellow) with the UAA stop codon (shown in stick representation). (d) Overall homology model of mtRF1a. (e) Average structure of mtRF1a (grey) after MD equilibration of the termination complex superimposed on the initial homology model (cyan). (f) Overall homology model of mtRF1. (g) Average structure of mtRF1 (grey) after MD equilibration of the termination complex superimposed on the initial homology model (magenta).
Modelling of mtRF1 with the included rRNA and the A1913C mutation resulted in a very reasonable structure where mtRF1 makes a hydrogen bond to the cytosine, similar to the interaction between Thr115 in RF1 and A1913 (Fig. 2). Arg193 of the RT insertion is predicted to adopt the position occupied by Arg298 in the so-called switch loop of RF1. Furthermore, in mtRF1 Arg298 corresponds to a glutamine and the switch loop is slightly shifted away, which increases the space for the RT loop to fit without disrupting the positioning of the rRNA. Thr194 of the insertion further stabilizes this conformation of the loop by hydrogen bonding to β5 (Fig. 2). In the recognition loop, the characteristic PxT motif corresponds to PKT in mtRF1a, whereas mtRF1 has a longer loop due to the GLS insertion. Here the loop in mtRF1 is predicted to adopt a conformation so that the inserted Ser169 aligns with the position of Thr186 in RF1 (see below), allowing the serine to bridge between the first and second codon positions of UAA as seen in crystal structures14. The homology model of mtRF1 actually shows more favourable interactions with the large subunit than mtRF1a, which has a valine at the position of Thr115 in RF1 and no polar contacts with the A1913C base.
Despite strong conservation among the three RFs in several regions, the sequence homology deteriorates distinctly upstream of the loop connecting domains 1 and 2, and the entire domain 1 differs considerably. It is clear that its contact regions with both the small and large ribosomal subunits are bound to be different in mtRF1. For example, there appears to be an insertion in the loop connecting helices α3 and α4 that would be likely to collide with rRNA helices 33 and 33a of the bacterial 30S subunit. Notably, these two rRNA helices are deleted in vertebrate mitoribosomes5. Similarly, the contact points between domain 1 and the 50S subunit involve sequences at the amino terminus of α3 and carboxy terminus of α4 where the RFs have very low homology. On the bacterial 50S subunit, these contact regions involve rRNA helices 43–44 and 95 to which the linker sequences have been shortened considerably in vertebrate mitoribosomes, which could imply a different positioning of the helices. Hence, shortening of the rRNA in the mitoribosome in regions distant from the decoding centre may well explain why mtRF1 shows no release activity with bacterial ribosomes.
To further explore the decoding characteristics of mtRF1a and mtRF1, MD simulations and free-energy calculations were performed with the homology models in the ribosomal A-site14, including the A1913C mutation. Here the initial MD simulations also serve as a refinement procedure that allows relaxation of the static models. In particular, it is necessary to evaluate the stability of the two loop insertions in mtRF1. As can be seen from Fig. 1e,g, the predicted structures of both mtRF1a and mtRF1 are remarkably stable during MD simulation with the template UAA codon in the ribosomal A-site and also have favourable backbone characteristics (Supplementary Fig. S1).
Energetics of stop codon reading
The MD free-energy calculations involve computing the relative binding free energies (ΔΔGbind) for the RFs between the cognate RF1 codons UAA and UAG, and the non-cognate codons UGA, AAA and AGA. For comparison, simulations of RF1 and RF2 binding to the bacterial ribosome were also carried out and the cognate-to-cognate UAA to UAG and UAA to UGA mutations for RF1 and RF2, respectively, thus serve as controls of cognate codon recognition. The calculated binding free energies are summarized in Fig. 3a (see also Supplementary Table S1), and provide a clear picture of the recognition pattern of the two mitochondrial RFs. As found earlier20, the calculated binding free energies for RF1 are very similar to those derived from experimental KM values21. Both mtRF1a and mtRF1 are predicted to behave qualitatively the same as RF1, with large discriminations towards both a first-position A and a second-position G. This is evident from both mutations of UAA into AAA and UGA, and the mutation of UGA into AGA. Moreover, the differences in binding affinity between UAA and UGA for mtRF1a and mtRF1 are large (+3.5 and +6.0 kcal mol−1, respectively) compared with RF2 (−0.6 kcal mol−1) for which UGA is cognate. Hence, there is no sign of either mtRF1a or mtRF1 being able to read the first two bases of the non-standard stop codons AGA and AGG. Just as for RF1, both mtRF1a and mtRF1 are found to be insensitive to A/G substitution at the third codon position, in accordance with the conservation of the key RF1 residues Gln181 and Thr194 (ref. 20).
(a) Calculated changes in binding free energy (kcal mol−1) for termination complexes with mtRF1a, mtRF1, RF1 and RF2 caused by different stop codon mutations. (b) Average interaction energies between the UAA codon bases and their surrounding environment for RF1 (yellow), mtRF1a (cyan), mtRF1 (magenta) and RF2 (green). Errors are given as the s.e.m. for ten independent simulations.
The question of whether mtRF1 has the ability to bind any stop codon at all is not addressed by the free-energy calculations in Fig. 3a, as they only yield binding free energies relative to UAA. However, by calculating the average MD interaction energies of the UAA codon for the four ribosome complexes with RF1, mtRF1a, mtRF1 and RF2 as done in earlier work on codon–anticodon recognition22, the absolute binding affinities of the four factors for UAA can be compared. This shows that the affinities in the decoding site are expected to be similar for all four release factors (Fig. 3b). Hence, although it is still beyond the scope of current simulation methods to predict absolute binding free energies to the entire ribosome, we can conclude that there is no indication of any major affinity differences among the different RFs for the messenger RNA codon.
Structural basis of mtRF1a and mtRF1 stop codon specificity
Besides yielding very similar interaction energies, the MD simulations with the UAA stop codon also show that the key interactions between the codon and the RFs are largely conserved (Fig. 4). Hence, just as in RF1 and RF2 the N terminus of the α5 helix with its conserved glycines reads the first-position U in both mtRF1a and mtRF1. Further, as noted above, Ser269 in the recognition loop of mtRF1 replaces the role of Thr186 in the RF1 PxT motif and recognizes both the first-position U and second-position A (Fig. 4c), which would not have been expected from the sequence alignment alone. The positioning of Ser269 in mtRF1 is consistently predicted both by the homology model and MD simulations.
(a–d) Average structures from MD simulations of RF1 (yellow), mtRF1a (cyan), mtRF1 (magenta) and RF2 (green) with key residues in stick representation and hydrogen bonds shown as dashed lines. The hydrogen bond network in the reading of UAA is similar for RF1, mtRF1a and mtRF1 in all three stop codon positions. Ser269 in mtRF1 interacts with U1 and A2 in a similar fashion as Thr206 in mtRF1a and Thr186 in RF1.
To further compare the structural features of stop codon reading, and possibly identify putative hotspots for AGA and AGG reading, average MD structures for mtRF1a and mtRF1 were analysed. Figure 5 shows the average structures from ten independent simulations of mtRF1a and mtRF1 with the UAA and AAA codons. With a first-position A, there is no unique stable conformation of the nucleotide, which is shifted away from the α5 helix disrupting the preceding loop structure for both of the mtRFs. In contrast, with a first-position U, the structures are remarkably stable (Fig. 5). Hence, it is evident for both mtRF1a and mtRF1 that there is too limited space for accommodating a purine at the first stop codon position owing to the location of the α5 helix. In particular, none of the mtRF1 insertion residues show any sign of favourable interactions with a first-position A, in contrast to the UAA codon, which shows the same strong hydrogen bonds with the α5 helix and the recognition loop that have previously been identified14,20.
(a) Average structures of ten independent MD simulations of mtRF1 (magenta) in complex with UAA (U1 in sticks), showing good structural convergence. (b) Average structures from the corresponding simulations with AAA show A1 shifted away from the tip of α5, with a less well-defined conformation. (c,d) Average structures from corresponding MD simulations of mtRF1a with the UAA and AAA codons, respectively.
The key residue enabling reading of a second-position G in RF2 is Glu128, the position of which is controlled by specific interaction network of mostly charged residues20,23. Although this residue is also a glutamate in RF1 (Glu119) and mtRF1a (Glu141), its interaction network is different in these RFs, prohibiting an efficient interaction with G2 as described earlier20. However, in mtRF1, the residue at this position is Ile199 and the entire cavity that would accommodate a second-position G is mostly hydrophobic. Hence, it is clear that Ile199 cannot contribute to a higher specificity for accommodating a G in the second position. This is illustrated in Fig. 6 where the average MD structures of RF2 and mtRF1 with the UGA codon are shown together with average G2 interaction energies with the binding site. mtRF1 shows no stabilization of a second-position G in contrast to RF2 where Glu128 is the key specificity element.
(a) Average interaction energies between G2 and the surrounding amino acids from MD simulations with the UGA codon show no stabilization of the second position G in mtRF1, in contrast to the case with bacterial RF2. Residue numbers are given in the format RF2/mtRF1, and green and magenta bars denote RF2 and mtRF1 energies, respectively. (b,c) Average structures of RF2 (green) and mtRF1 (magenta) zoomed in on the second stop codon position. Ile199 is unable to form the hydrogen bonds with G2 seen in the RF2 UGA complex.
Apart from the insertion residues in the recognition loop, mtRF1 shows a difference in the positioning of Arg274 compared with the corresponding Arg residue in RF1, RF2 and mtRF1a. This appears to be caused by the absence of the negatively charged residue found in the other RFs (Glu187 in RF1, Asp196 in RF2 and Glu209 in mtRF1a), which in mtRF1 allows Arg274 to form strong interactions with the mRNA backbone of the first stop codon position and the second position base (Supplementary Fig. S2). In addition, Arg274 makes hydrogen bonds with both Ser269 and His276, as well as with the sugar backbone of the P-site mRNA via a bridging water molecule. This extensive hydrogen bond network of Arg274 thus contributes to stabilization of the considerably larger recognition loop in mtRF1.
An alternative mechanism for AGA and AGG termination
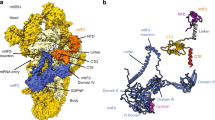
To investigate the reported hydrolysis induced by ICT1 at the non-standard stop codons9, a homology model of ICT1 was constructed based on the structure of the bacterial homologue YaeJ. YaeJ is a rescue factor for stalled ribosomes and its ribosome complex was solved with a short mRNA ending in the P-site, as would be the case for stalled ribosomes with an empty A-site24. In this structure, the C-terminal tail of YaeJ is positioned in the mRNA channel, where it is able to detect the missing A-site codon. Moreover, YaeJ has been shown to rescue ribosomes stalled due to rare AGG codon clusters25. Interestingly, the path of the C-terminal tail of YaeJ observed in the crystal structure24 would not be possible with a codon in the A-site, as the tail would clash with the mRNA. Moreover, the observed peptidyl-tRNA hydrolysis caused by ICT1 with several codons (including AGA and AGG)9 therefore suggests that the crystal structure path of YaeJ24 is not compatible with ICT1 binding with an A-site codon present. However, two alternative paths of YaeJ can, in fact, be identified and Fig. 7a shows these conformations together with the crystal structure of YaeJ, aligned to a ribosome with A-site mRNA present26.
(a) Two possible paths for YaeJ (green and cyan) with mRNA (blue) present in the mRNA channel. The A-site codon in highlighted in red and the P-site tRNA is shown in orange. The yellow structure corresponds to the known path of YaeJ on a ribosome without an A-site codon and with an empty mRNA channel. (b) Homology model of ICT1 (magenta) positioned in one of the possible paths. The insertion in ICT1 and four additional amino acids were modelled as an extension (dark green) of the α-helix according to the secondary structure prediction. (c) Alignment and secondary structure prediction of the C-terminal end or ICT1 and YaeJ.
Sequence alignment of human ICT1 and E. coli YaeJ also reveals an insertion of six residues in ICT1 preceding the α-helix at the end of YaeJ. This insertion is consistently assigned by secondary structure prediction tools27,28 to be a part of the C-terminal helix (Fig. 7c). The homology model of ICT1 was created based on one of the alternative paths of YaeJ as template. The insertion predicted to be part of the helix is then positioned close to the A-site codon (Fig. 7b). Similar to YaeJ, this part of the helix has several positively charged residues that could interact with the phosphate backbone of the mitoribosome and possibly the mRNA phosphates, thereby providing a termination mechanism for ribosomes stalled at the AGA and AGG codons.
Discussion
The conclusion from our extensive MD free-energy calculations on the homology models of mtRF1a and mtRF1 is that neither of these RFs is able to cause termination at the non-standard stop codons. Instead, we find that both mtRF1a and mtRF1 behave very much like the bacterial RF1, both with regard to codon specificity and interactions of key residues involved in codon binding20. The ability to read a first-position purine, as would be required for the AGA and AGG codons, is essentially prevented by the position of the α5 helix, which is highly unlikely to differ between the mtRFs and RF1. Even though the bacterial and the mitochondrial ribosomes are bound to be rather different overall, the fact that the decoding region of the mitochondrial 9S and the bacterial 16S RNA are conserved in secondary structure4,29 strongly suggests that this also pertains on the local three-dimensional structure. This is also supported by the strong sequence conservation between RF1 and mtRF1a in the decoding region, which, together with the fact that the experimentally observed specificity of mtRF1a on bacterial ribosomes11,12, is reproduced here, strengthens the reliability of our derived homology models.
One of the two main hypotheses regarding termination at non-standard stop codons suggested a mechanism based on a −1 frameshift17. This would in human mitochondria result in the standard UAG stop codon, as both CO1 and ND6 has a U preceding AGA and AGG, respectively. This experimental evidence for this theory relies entirely on the capability of RelE to only cut the mRNA between the second and third position, which can, however, be questioned, as RelE has shown activity at different positions and codons in vivo30. Furthermore, it has been pointed out that human mitochondria are not the only vertebrate ones using non-standard stop codons13. Bioinformatics investigations have, in fact, shown that a U preceding AGA and AGG is rare31 and Supplementary Table S2 shows a few examples of vertebrates using non-standard stop codons, where most of the preceding codons end with an A. The second hypothesis suggests that mtRF1 is the RF causing termination on AGA and AGG. This was mainly based on the ability of a hybrid RF1, which was mutated to resemble mtRF1, to show a rather weak hydrolysis activity at the AAG (and UAA) codon13. However, its activity on AGA and AGG was barely above the background with no codon. It would thus seem that the hybrid RF1 has lost both activity and specificity by the mutations introduced, rather than obtaining a possible specificity for an A in the first position. Furthermore, it is clear that there are correlated mutations across the chimeric boundaries used in ref. 13. For example, the α5 RT insertion is correlated with mutations in the switch loop (see above) in addition to the A1913C rRNA mutation, which explains why the hybrid that only included the RT-insertion region from mtRF1 was completely inactive13.
As our calculations show no indication of mtRF1a or mtRF1 acting on the non-standard stop codons, this raises the key question whether there is any other possible mechanism for the termination at AGA and AGG. Here the two other members of the mitochondrial RF family, ICT1 and C12orf65, are of considerable interest, as they have been shown to be vital for mitochondrial translation. In fact, the only member of the mtRF family that has shown any in vitro activity on AGA and AGG is ICT1 (ref. 9). A mechanism where the mitoribosome becomes stalled at AGA or AGG, which lack cognate tRNAs, and then rescued by ICT1 would provide a possible explanation for the existence of non-standard stop codons. The homology model of ICT1 presented here reveals an extension of the α-helix, which in the YaeJ–ribosome complex24 is located at the A-site codon position. This helical structure could potentially assist in sensing a codon for which there is no tRNA and cause termination.
Our calculated energetics show that mtRF1a and mtRF1 have overlapping specificities for the UAA and UAG stop codons, comparable to the overlapping specificity for UAA by RF1 and RF2 in bacteria. This could be one reason for the limited effect on protein levels reported on mtRF1a depletion12. It is also a distinct possibility that mtRF1 is, in fact, the major release factor in vertebrate mitochondria, as mtRF1 has apparently not been tested in knockdown experiments. In this respect, measurements of release activity on bacterial ribosomes11,12 may be less relevant for the activity on the mitoribosome. It should also be mentioned that earlier homology modelling suggested that mtRF1 cannot read any codon but recognizes ribosomes with an empty A-site32. However, the mtRF1 homology model was in that case based on bacterial RF structures with no rRNA or mRNA included32, which may drastically affect the conformational space accessible to the protein. Structural relaxation by MD simulations and free-energy calculations further provide critical tests of the stability and energetics associated with homology models and are essential for drawing conclusions regarding binding properties. What speaks in favour of mtRF1 being the major release factor is the fact that the emergence of mtRF1 in vertebrates appears to be correlated with major changes in the mt-rRNA. That is, the consistent shortening of the mt-rRNA in vertebrates may have required concomitant adaptation of a release factor to be effective on these mitoribosomes. mtRF1 could thus be a factor that has specifically co-evolved with the vertebrate mitoribosome to adapt to its new rRNA structure. Another observation that may support a key role for mtRF1 is that its discrimination against UGA (which codes for Trp in vertebrate mitochondria) is predicted here to be significantly stronger than that of mtRF1a. As noted above, the key mutation to a hydrophobic residue (Ile199) near the second codon position strongly prevents UGA reading, suggesting that mtRF1 is evolved for the recoding of this codon in mitochondria.
Methods
Homology modelling
Homology models of mtRF1a and mtRF1 were created using Prime 2.1 (Schrödinger, LLC, New York, NY, 2007) and Modeller (Version 9.10)33 with the bacterial RF1–ribosome complex14 as template, including nearby rRNA, the stop codon and the A1913C mutation. The homology model of ICT1 was created with Prime 2.1 and the crystal structure of YaeJ–ribosome complex24 as template. The alignment was further compared with the solution structure of the Mus musculus ICT1 (ref. 34) to get the correct secondary structure alignment. The helix close to the C-terminal end (missing in ref. 34) was modelled as predicted by PSIPRED28 (Fig. 7). The model was minimized in MacroModel 9.5 (Schrödinger, LLC) using the OPLS-AA force field35.
Molecular dynamics and free-energy simulations
Relative binding free energies (ΔΔGbind) between different termination complexes were calculated using the free-energy perturbation (FEP) technique (see, for example, Brandsdal et al.36) and a standard thermodynamic cycle20. Initial coordinates for the MD and FEP calculations were taken from the crystal structure of RF1 and RF2 bound to the 70S ribosome14,23 and the corresponding homology models of mtRF1 and mtRF1a. For the mtRF1a and mtRF1 structures, A1913 of the 70S ribosome was changed to a C in accordance with the human mitoribosome sequence. The RF histidine residue between the second and third codon position was treated as neutral and protonated at ND1 (as there is an arginine residue close to NE2), and magnesium ion positions were selected from previous calculations20. To evaluate the changes in binding free energy, a set of different mRNA codon transformations (mutations) were carried out during MD simulations with and without the relevant release factor bound to the solvated ribosome–mRNA complex.
The MD/FEP calculations were carried out with spherical systems (40 Å in diameter) centred on the nucleotide base that was transformed (see Fig. 3 for the different transformations). All MD simulations were carried out with the Q programme37 using the Charmm22 force field38,39. The simulations were done at 300 K after stepwise heating of the system from 1 K. Water molecules close to the sphere boundary were subjected to radial and polarization restraints according the SCAAS model37,40. The MD simulations were carried out with a time step of 1 fs, and solvent bonds and angles were treated with the SHAKE algorithm41. Non-bonded interactions were treated with a 10-Å direct cutoff, and long-range electrostatic interactions beyond this cutoff were treated with the local reaction field method42. No cutoffs were applied to the transformed bases, and charged groups close to the sphere boundary were neutralized as described elsewhere43. Ten independent FEP simulations of each system were performed using different initial conditions. All FEP calculations were carried out using 51 windows (λ-points) interspaced for optimal efficiency with small λ-steps (0.0008) near the end points, with 200 ps of simulation time for each window36, resulting in a total data collection time of 100 ns after equilibration and heating. For all first-position calculations transforming U into A, the dual topology method was used, owing to the larger changes involved when transforming a pyrimidine into a purine, whereas the single topology method was used for A-to-G mutations.
Additional information
How to cite this article: Lind, C. et al. Codon-reading specificities of mitochondrial release factors and translation termination at non-standard stop codons. Nat. Commun. 4:2940 doi: 10.1038/ncomms3940 (2013).
References
McLean, J. R., Cohn, G. L., Brandt, I. K. & Simpson, A. M. Incorporation of labeled amino acids into the protein of muscle and liver mitochondria. J. Biol. Chem. 233, 657–663 (1958).
Anderson, S. et al. Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (1981).
Koc, E. C., Haque, M. E. & Spremulli, L. L. Current views of the structure of the mammalian mitochondrial ribosome. Isr. J. Chem. 50, 45–59 (2010).
Sharma, M. R., Booth, T. M., Simpson, L., Maslov, D. A. & Agrawal, R. K. Structure of a mitochondrial ribosome with minimal RNA. Proc. Natl Acad. Sci. USA 106, 9637–9642 (2009).
Sharma, M. R. et al. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115, 97–108 (2003).
O'Brien, T. W. Properties of human mitochondrial ribosomes. IUBMB Life 55, 505–513 (2003).
Sengupta, S., Yang, X. & Higgs, P. G. The mechanisms of codon reassignments in mitochondrial genetic codes. J. Mol. Evol. 64, 662–688 (2007).
Jukes, T. H. & Osawa, S. Evolutionary changes in the genetic code. Comp. Biochem. Physiol. B 106, 489–494 (1993).
Richter, R. et al. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 29, 1116–1125 (2010).
Zhang, Y. & Spremulli, L. L. Identification and cloning of human mitochondrial translational release factor 1 and the ribosome recycling factor. Biochim. Biophys. Acta 1443, 245–250 (1998).
Nozaki, Y., Matsunaga, N., Ishizawa, T., Ueda, T. & Takeuchi, N. HMRF1L is a human mitochondrial translation release factor involved in the decoding of the termination codons UAA and UAG. Genes Cells 13, 429–438 (2008).
Soleimanpour-Lichaei, H. R. et al. mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol. Cell 27, 745–757 (2007).
Young, D. J. et al. Bioinformatic, structural, and functional analyses support release factor-like MTRF1 as a protein able to decode nonstandard stop codons beginning with adenine in vertebrate mitochondria. RNA 16, 1146–1155 (2010).
Laurberg, M. et al. Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857 (2008).
Antonicka, H. et al. Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect. Am. J. Hum. Genet. 87, 115–122 (2010).
Kogure, H. et al. Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65. Proteins 80, 2629–2642 (2012).
Temperley, R., Richter, R., Dennerlein, S., Lightowlers, R. N. & Chrzanowska-Lightowlers, Z. M. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science 327, 301–301 (2010).
Åqvist, J., Lind, C., Sund, J. & Wallin, G. Bridging the gap between ribosome structure and biochemistry by mechanistic computations. Curr. Opin. Struct. Biol. 22, 815–823 (2012).
Trobro, S. & Åqvist, J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol. Cell 27, 758–766 (2007).
Sund, J., Andér, M. & Åqvist, J. Principles of stop-codon reading on the ribosome. Nature 465, 947–950 (2010).
Freistroffer, D. V., Kwiatkowski, M., Buckingham, R. H. & Ehrenberg, M. The accuracy of codon recognition by polypeptide release factors. Proc. Natl Acad. Sci. USA 97, 2046–2051 (2000).
Almlöf, M., Andér, M. & Åqvist, J. Energetics of codon-anticodon recognition on the small ribosomal subunit. Biochemistry 46, 200–209 (2007).
Korostelev, A. et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl Acad. Sci. USA 105, 19684–19689 (2008).
Gagnon, M. G., Seetharaman, S. V., Bulkley, D. & Steitz, T. A. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science 335, 1370–1372 (2012).
Handa, Y., Inaho, N. & Nameki, N. YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res. 39, 1739–1748 (2011).
Jenner, L. B., Demeshkina, N., Yusupova, G. & Yusupov, M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560 (2010).
Cole, C., Barber, J. D. & Barton, G. J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 (2008).
Jones, D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999).
de la Cruz, V. F., Lake, J. A., Simpson, A. M. & Simpson, L. A minimal ribosomal RNA: sequence and secondary structure of the 9S kinetoplast ribosomal RNA from Leishmania tarentolae. Proc. Natl Acad. Sci. USA 82, 1401–1405 (1985).
Hurley, J. M., Cruz, J. W., Ouyang, M. & Woychik, N. A. Bacterial toxin RelE mediates frequent codon-independent mRNA cleavage from 5' end of coding regions in vivo. J. Biol. Chem. 286, 14770–14778 (2011).
Duarte, I., Nabuurs, S. B., Magno, R. & Huynen, M. Evolution and diversification of the organellar release factor family. Mol. Biol. Evol. 29, 3497–3512 (2012).
Huynen, M. A., Duarte, I., Chrzanowska-Lightowlers, Z. M. A. & Nabuurs, S. B. Structure based hypothesis of a mitochondrial ribosome rescue mechanism. Biol. Direct 7, 14 (2012).
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Handa, Y. et al. Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality. J. Mol. Biol. 404, 260–273 (2010).
Jorgensen, W. L., Maxwell, D. S. & Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 (1996).
Brandsdal, B. O. et al. Free energy calculations and ligand binding. Adv. Protein Chem. 66, 123–158 (2003).
Marelius, J., Kolmodin, K., Feierberg, I. & Åqvist, J. Q. A molecular dynamics program for free energy calculations and empirical valence bond simulations in biomolecular systems. J. Mol. Graph. Model. 16, 213–225 (1998).
Mackerell, A. D. Jr, Wiorkiewicz-Kuczera, J. & Karplus, M. An all-atom empirical energy function for the simulation of nucleic acids. J. Am. Chem. Soc. 117, 11946–11975 (1995).
MacKerell, A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998).
King, G. & Warshel, A. A surface constrained all-atom solvent model for effective simulations of polar solutions. J. Chem. Phys. 91, 3647–3661 (1989).
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Lee, F. S. & Warshel, A. A local reaction field method for fast evaluation of long-range electrostatic interactions in molecular simulations. J. Chem. Phys. 97, 3100–3107 (1992).
Trobro, S. & Åqvist, J. Mechanism of peptide bond synthesis on the ribosome. Proc. Natl Acad. Sci. USA 102, 12395–12400 (2005).
Acknowledgements
Support from the Swedish Research Council (VR), the Knut and Alice Wallenberg Foundation and the Swedish National Infrastructure for Computing (SNIC) is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
C.L. and J.S. performed experiments. J.Å. designed experiments. C.L., J.S. and J.Å. analysed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1-S2, Supplementary Tables S1-S2 and Supplementary Reference (PDF 396 kb)
Rights and permissions
About this article
Cite this article
Lind, C., Sund, J. & Åqvist, J. Codon-reading specificities of mitochondrial release factors and translation termination at non-standard stop codons. Nat Commun 4, 2940 (2013). https://doi.org/10.1038/ncomms3940
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms3940
This article is cited by
-
Human mitochondria require mtRF1 for translation termination at non-canonical stop codons
Nature Communications (2023)
-
Mitochondria homeostasis: Biology and involvement in hepatic steatosis to NASH
Acta Pharmacologica Sinica (2022)
-
Human mtRF1 terminates COX1 translation and its ablation induces mitochondrial ribosome-associated quality control
Nature Communications (2022)
-
Mechanisms and regulation of protein synthesis in mitochondria
Nature Reviews Molecular Cell Biology (2021)
-
Mechanism of ribosome rescue by alternative ribosome-rescue factor B
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.