Abstract
Bacterial persistence has been shown to be an underlying factor in the failure of antibiotic treatments. Although many pathways, among them the stringent response and toxin–antitoxin modules, have been linked to antibiotic persistence, a clear molecular mechanism for the growth arrest that characterizes persistent bacteria remained elusive. Here, we screened an expression library for putative targets of HipA, the first toxin linked to persistence, and a serine/threonine kinase. We found that the expression of GltX, the glutamyl-tRNA-synthetase, reverses the toxicity of HipA and prevents persister formation. We show that upon HipA expression, GltX undergoes phosphorylation at Ser239, its ATP-binding site. This phosphorylation leads to accumulation of uncharged tRNAGlu in the cell, which results in the activation of the stringent response. Our findings demonstrate a mechanism for persister formation by the hipBA toxin–antitoxin module and provide an explanation for the long-observed connection between persistence and the stringent response.
Similar content being viewed by others
Introduction
The phenomenon of bacterial persistence to antibiotics has been extensively studied in the past decade1,2. Persistence is observed when a fraction of a clonal bacterial population survives extensive antibiotic treatments that have efficiently killed off the majority of the population3. In contrast to resistance, persistence is a transient phenotype and not linked to genetic alterations4. Persistence can be viewed as a powerful survival strategy of bacteria within a genetically uniform population to cope with unpredictable stress1,2 through the stochastic formation of a small subpopulation of transiently dormant bacteria refractive to many antibiotics that target actively growing cells, as well as to phages5 and temperature6. Although other phenotypes have been linked to persistence, dormancy is the most commonly reported survival factor1. Several toxin–antitoxin (TA) modules have been linked to the persistent phenotype7,8, with hipAB being the first one directly correlated to this phenotype9. Although understanding of the environmental conditions needed to trigger persistence through TA modules is still lacking, the hipBA module and its high-persistence mutant, hipA7, constitute a ‘model system’ for studying persistence that appears upon starvation, namely Type I persistence. HipA is a toxin and a serine/threonine kinase that is capable of auto-phosphorylation10 and the phosphorylation of EF-Tu11, causing growth arrest and persister formation when the HipA level is higher than HipB. Ectopic expression of HipA arrests cell growth and mediates the formation of persister cells12. Using this system, we have previously shown that the key feature of persistence, namely the coexistence of dormant and actively growing cells, is due to a threshold mechanism resulting from the titration of HipA by HipB13. This threshold is a general feature of TA modules. However, the molecular mechanism through which HipA arrests growth and controls the duration of the growth arrest remains unclear. The observation that mutants in the stringent response pathway create less persisters than the wild-type Escherichia coli cells14,15 suggests that the stringent response plays an important role in persister formation16,17. In agreement with this understanding, activation of the stringent response by HipA has recently been reported18. The absence of induction of the stringent response upon inhibition of EF-Tu19 suggests that the phosphorylation of EF-Tu may be only part of the explanation for the growth arrest observed upon hipA expression.
Our goal was to further investigate the connection between HipA, growth arrest and the stringent response. We found that HipA activates the stringent response by phosphorylation of GltX, which is an aminoacyl tRNAGlu synthetase, at its ATP-binding site. This phosphorylation inactivates GltX, leads to increased levels of uncharged tRNAGlu, thus activating RelA, and increases (p)ppGpp levels.
Results
HipA7 cold sensitivity phenotype is suppressed by GltX
In order to search for the mediating mechanism between HipA expression and (p)ppGpp induction, we took advantage of the MG1655A7 (Supplementary Table S1) mutant9. Two point mutations were found in hipA7 (ref. 14) that trigger high persistence and cold sensitivity6,9. Using the cold-sensitive phenotype for selection, we transformed MG1655A7 cells with a genomic E. coli library20. The purified plasmids, which were collected from the colonies that overcame the cold-sensitive phenotype, were found to be in two groups: plasmids containing the hipAB operon together with part of the yneO gene, which is a pseudogene (10 clones) and plasmids containing the gltX gene (9 clones), a glutamyl-tRNA-synthetase, together with YfeD, which is predicted to be an antitoxin from an uncharacterized TA system (Supplementary Fig. S1). We cloned the gltX gene separately and found that MG1655A7 bacteria overexpressing GltX were the only one able to grow at 20 °C.
GltX prevents ppGpp synthesis upon HipA overexpression
In order to test whether the growth arrest caused by HipA is directly related to the activation of the stringent response, we transformed hipB+ cells (MGNR— Supplementary Table S1) with a plasmid carrying pTet-hipA-mcherry13, induced the overexpression of HipA and monitored the levels of ppGpp and pppGpp (collectively named (p)ppGpp) which are the hallmarks of stringent response activation21. Clear (p)ppGpp synthesis was observed 30 min after the induction of HipA in agreement with a recent report18 (Fig. 1a), indicating that HipA expression activates the stringent response.
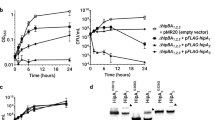
MGNR cells overexpressing HipA were grown in 100 μl medium (as described in the Methods section). Samples of 10 μl were separated by thin-layer chromatography. Representative autoradiogram of PEI thin-layer chromatography of the accumulation of (p)ppGpp in MGNR cells overexpressing HipA with different levels of GltX: (a) GltX+ cells. (b) GltX++ cells. (c) GltXwt cells carrying pQE30 empty vector (d) GltXwt cells carrying pQE30 empty vector with the addition of IPTG. (e) Quantification of a–d. a- Red; b- Yellow; c- Green; d- Purple. Error bars represent s.d. of duplicates from two different experiments.
In order to determine whether GltX prevents the activation of the stringent response that is induced by HipA, we added another plasmid controlling the level of gltX (pTac-gltX) (Supplementary Table S2). The Tac promoter is known to be leaky, resulting in three levels of GltX expression: (a) ‘GltXwt’: expression from the native chromosomal gene; (b) ‘GltX+’:GltXwt together with leaky expression from pTac-gltX; and (c) ‘GltX++’:GltXwt together with maximal induction from pTac-gltX. We monitored (p)ppGpp synthesis after the induction of HipA in the presence (Fig. 1b) and absence (Fig. 1a,c,d) of overexpressed GltX. We found that (p)ppGpp synthesis was completely abolished in GltX++ cells despite the overexpression of HipA. The inhibition of (p)ppGpp synthesis upon HipA induction by GltX is dependent on the levels of HipA. In the present experiment, the levels of HipA were maximal (see Materials and Methods), and thus, the leakiness from pGltX (GltX+) is not sufficient to completely reduce (p)ppGpp synthesis (Fig. 1a,e). Maximal levels of GltX abolished the effect of HipA on (p)ppGpp synthesis (Fig. 1b,e).
GltX prevents growth arrest mediated by HipA
We hypothesized that the prevention of (p)ppGpp synthesis by GltX overexpression would also abolish the growth arrest caused by HipA, as well as persister formation. We first measured the effect of co-expressing GltX on growth upon HipA overexpression with anhydrotetracycline (aTc). GltX+ cells grew normally despite the overexpression of HipA, whereas GltXwt cells were growth arrested (Fig. 2a,b). We previously showed that the growth arrest inflicted by HipA is transient and results in an extended lag time. The expression level of HipA above a threshold determines the duration of the lag time and thus the level of persistence13. We measured the effect of GltX on the persistence induced by HipA by monitoring the lag time distribution by the ScanLag method22, with and without overexpression of GltX. As previously reported, cells expressing HipA above the threshold exhibited an extended lag with a broad distribution of colony appearance—a ‘tail’ which represents the persisters subpopulation. In contrast, GltX+ cells expressing low and medium levels of HipA did not exhibit the extended growth arrest. Only at very high levels of HipA expression the lag time was again extended despite the presence of extra GltX (Fig. 2c,d). Thus, GltX prevents the formation of growth-arrested cells caused by the accumulation of HipA.
(a) Growth curve of hipB+ cells carrying the HipA plasmid only. (b) Growth curve of hipB+ cells carrying HipA plasmid together with pGltX. The optical density was measured in a multiplate reader (Victor2, Perkin-Elmer) every 2.5 min. Statistical information is shown in Supplementary Fig. S5. (c,d) Cells from a or b, respectively, were plated on LB 3 h after aTc addition. The appearance time of colonies was continuously monitored by the ScanLag system. The histograms show the fraction of CFUs detected at each time point. (e) Survival curve of cells under Norfloxacin treatment. Solid lines—cells carrying pHipA and pQE30 (GltXwt cells) without IPTG addition; Dashed lines—cells carrying pHipA and pGltX (GltX+ cells). Blue—without the addition of ATC; Red—with 4 ng ml−1 aTc. Four independent assays show significant lower persistence in GltX+ conditions, with a P-value of 0.017, using the t-test. Error bars represent s.d. of duplicates from three different experiments.
GltX reduces antibiotic persister levels
The above described phenotype implies that GltX should also reduce persistence to antibiotic treatments. To test this prediction, we monitored the survival under Norfloxacin treatment of cultures overexpressing HipA, with and without the overexpression of GltX (Fig. 2e). We found that the high-survival level acquired by cultures overexpressing HipA disappeared in GltX+ cells. These results show that increased levels of GltX preclude the onset of the stringent response activated by HipA, preventing growth arrest and persister formation.
GltX is phosphorylated upon induction of HipA
A common method to induce the stringent response is by inhibiting tRNA-synthetases23, suggesting that HipA may inhibit GltX. HipA was shown to be a serine/threonine kinase capable of auto-phosphorylation10 and the phosphorylation of other targets in the cell11. In order to determine whether GltX is phosphorylated as a result of HipA overexpression, cell cultures containing plasmids overexpressing both HipA and GltX were grown in the presence of 32P and the labelled protein profile analyzed by gel electrophoresis. A band the size of GltX (56 kDa) can be seen in the whole-cell extract only upon HipA overexpression (Fig. 3a). Stronger support for the phosphorylation of GltX in the presence of high HipA levels is found in purified his-GltX. A clear radioactive band was seen in the purified fraction at the size of 56 kDa, again only in cells that overexpressed HipA (Fig. 3b). We verified that his-GltX was equally expressed in both cultures, namely in cells that did and did not express HipA, by western blot analysis to the same membrane as in Fig. 5b (Fig. 3c). Using Mass Spectrometry (MS) analysis, we confirmed that GltX is phosphorylated upon HipA overexpression (Fig. 3d,e). Interestingly, we found by MS-MS that the phosphorylation site of GltX is a serine (Ser239) (Fig. 3f and Supplementary Fig. S2). This residue is part of the KKLSKR motif in the ATP-binding site of GltX24, suggesting that HipA may inactivate GltX by phosphorylation and is in agreement with predictions by Sekine et al.24, that a mutation at this site would change the conformation of the ATP-binding site thus leading to GltX inactivation. As further corroboration with this prediction, we show that the induction of HipA leads to the accumulation of uncharged tRNAGlu, whereas overexpression of GltX (GltX++) prevents this accumulation (Supplementary Fig. S3).
(a–c) hipB+ cells carrying two plasmids expressing HipA and his-GltX grown in the presence of 32P. All cells were GltX++. (−) refers to cells that do not express HipA; (+) refers to cells overexpressing HipA by the addition of aTc. (a) Whole-cell extract was separated by electrophoresis, transferred to a PVDF membrane and exposed to a hyper sensitive film. (b) His-GltX was separated from the whole-cell extract by Ni-bids, further treated as in (a). (c) Western blot analysis of the membrane in b using an anti-his antibody. (d,e) MS analysis MS of purified GltX as described in b. (d) MS analysis of GltX without the induction of HipA. (e) MS analysis of GltX with the induction of HipA. Red box marks the location of the peptide containing the phosphorylated serine. (f) MS-MS analysis of the phosphorylated peptide of GltX, by the Protein Discoverer 1.3 software, using a cutoff of 0.8 Da. The table and the spectrum show the ‘y’ singly charged ions of the phosphorylated peptide. In the spectrum the distance between the peaks represent the (m/z) of each amino acid. The spectra of the whole phosphorylated and non-phosphorylated peptides are shown in Supplementary Fig. S2.
Discussion
Our findings as well as similar results obtained while this work was under revision25 can serve to fill in a missing link in the cycle describing the formation of persisters by the hipBA TA module (Fig. 4). Overexpression of HipA inactivates GltX by phosphorylating its ATP-binding site, leaving GltX unable to covalently link the glutamate amino acids to their cognate tRNAGlu (Supplementary Fig. S3). Increased levels of uncharged tRNAGlu in the cells trigger RelA to synthesize (p)ppGpp, activating the stringent response26 and thus leading to dormancy and persistence. This pathway now closes the loop of persister formation when combined with the results of an earlier model27, suggesting that (p)ppGpp is the regulator of persistence by activating TA loci28 via PolyP and Lon29,30, which has been shown to degrade HipB31. We had shown that a triggering mechanism leading to an imbalance of activation of HipA toxicity is needed to bring the bacterial population closer to the threshold and generate Type I persisters. Typically, Type I persistence is triggered by starvation conditions. When MG1655A7 cells are maintained at strictly exponential growth, persistence vanishes10,32. We propose that an initial activation of the stringent response by starvation increases the level of (p)ppGpp, and activates the persistence cycle in the cells in which free HipA is near the threshold (Fig. 4). Free HipA causes the phosphorylation of GltX, further triggering the stringent response due to the presence of uncharged tRNAs as well as increasing cellular (p)ppGpp levels26. The interplay between the stringent response and the action of the HipA toxin suggests new ways to target persister cells by interfering directly with the stringent response. Indeed, the use of novel analogues of ppGpp designed to inhibit RelA enzymes that interfere with long-term bacterial survival strategies33 may thus present a promising candidate for new specific anti-persister therapies34.
(1) Increased levels of ppGpp at the entrance to stationary phase lead to accumulation of inorganic polyphosphate (polyP)29. (2) PolyP promotes Lon degradation activity27,30; (3) HipB, is degraded by Lon31 leading to (4) free active HipA. (5) Free HipA inactivates GltX by phosphorylation of its ATP-binding site (ser239). (6) Phosphorylated GltX is unable to charge tRNAGlu with glutamate, leading to increased levels of uncharged tRNAGlu. (7) RelA is activated by uncharged tRNAGlu to synthesize more (p)ppGpp, closing the cycle and (8) leading to growth arrest and persister formation. Note that HipA action on GltX is probably only one of the pathways closing the cycle.
The persistence cycle activates the stringent response, together with growth arrest, in a runaway process that further sharpens the threshold separating persisters from normal cells. We have added the positive feedback of the free HipA level on the degradation rate of HipB, via the inactivation of GltX, to our previous model of the hipBA TA module describing the persistence level as a function of the total toxin level in cells, AT (ref. 13) (Supplementary Fig. S4). The analytic solution of the model (Supplementary Table S3) now predicts the level of uncharged tRNAGlu, a proxy for the persistence level, as a function of the levels of HipA and GltX and reproduces the reduction in persistence that we observed upon GltX overexpression (Supplementary Fig. S4). Note that while the HipA–GltX interaction represents the first closure of the persistence cycle, it is likely, in light of the identical persistence level of hipA deletion mutant35,36, that several other parallel ways may close the cycle in a similar way.
The model proposed in Fig. 4 describes the induction of Type I persistence by any step that may start the persistence cycle, in particular starvation and activation of the stringent response. Type I persistence triggered by starvation is the most common form of persistence1. For example, it was shown recently in Pseudomonas aeruginosa that the response to starvation actively mediates antibiotic tolerance and biofilm formation15. Our results show that the mutual activation of the stringent response and the hipBA TA locus can provide a mechanism for Type I persistence, by inactivation of aminocyl-tRNA synthetases, a mechanism which may be used by other TA modules in parallel persister activation cascades37 that converge on the activation of the stringent response. This is consistent with the observation that Type I persistence decreases in cells impaired for the stringent response both in planktonic cells14,38,39 and in biofilms15. The novel HipA target revealed in our study therefore serves to uncover another piece in the puzzle of HipA-mediated persistence.
Methods
Media and reagents
Bacterial strains used in this study are described in Supplementary Table S1. Plasmid construction is described in Supplementary Table S2. All E. coli strains were grown at 37 °C in LB medium unless indicated differently. The antibiotics used for selections were used at the following concentrations: ampicillin 100 μg ml−1, kanamycin 50 μg ml−1, tetracycline 20 μg ml−1, and chloramphenicol 34 μg ml−1.
In vivo (p)ppGpp accumulation assay
ΔhipAhipB+ (MGNR) cells carrying pZA21HipA-mcherry plasmid13 and pGltX plasmid, as indicated in the text, were grown in MOPS glucose minimal medium40 supplemented with all amino acids. At OD600 0.1, cells were labelled with H332PO4, and 1.2 μg ml−1 of isopropylthiogalactoside (IPTG) was added when necessary for the induction of GltX and incubated for 1 h, after which HipA was induced by the addition of 50 ng ml−1 of aTc. Samples were withdrawn 30 and 60 min after addition of aTc. Reactions were stopped by the addition of 25 μl of formic acid per 100 μl reaction volume. Aliquots (10 μl) of each reaction were loaded and separated on Cellulose PEI TLC plates (Merck) using 1.5 M KH2PO4 as mobile phase. Plates were autoradiographed using the Fijix Bas100 Phosphor Imager (Japan). (p)ppGpp signal was measured using TINA 2.0 software (Raytest, Strauben-Hardt).
Library screen
MG1655A7 cells were transformed with a genomic E. coli library mix20. Cells were plated on ampicillin plates and incubated at 20 °C for 48 h. As controls, we used the MGH strain that did not contain any plasmid, and the MGH strain containing an ample pBR vector. Plasmids were purified from the colonies that succeeded to grow at 20 °C. MGH cells were transformed again, this time with each of the purified plasmids and incubated at 20 °C for 48 h to verify that the phenotype is restored. Plasmids capable of restoring the cold sensitivity phenotype were sequenced.
Cloning of genes from the library screen
The GltX and YfeD genes were cloned separately into the pQE30 vector under a his-tag with the primers:
FW-5′-ACGAGAGCTCATGAAAATCAAAACTCGCTT-3′,
REV-5′-ATATGTCGACTTACTGCTGATTTTCGCGTT-3′,
FW-5′-ATACGAGCTCATGAAAAGATTACGCAATAA-3′ and
REV-5′-TTAAGTCGACTTATGCTGATTCGTCAATAT-3′, respectively. As templates, we used the plasmids purified from the library screen.
Protein preparation and analysis
MGNR cells carrying relevant plasmids were grown in MOPS glucose minimal medium40 supplemented with all amino acids. At OD600 0.1, cells were labelled with H332PO4 and 1.2 μg ml−1 of IPTG was added for the induction of GltX when necessary. After 1 h of incubation, HipA expression was induced by the addition of 50 ng ml−1 of aTc. After another 45 min, cells were harvested by centrifugation and lysed by resuspension in a BugBuster solution (Novagen). His-tagged GltX was purified from the lysate, as described before33. For analysis of the protein samples from whole cells, lysate and purified GltX were separated by 12% SDS-polyacrylamide gel electrophoresis and transferred to PVDF membrane (Millipore Bedford). The membrane was autoradiographed by using Hyper Sensitive films (GE Health Care) for 4 h. The same membrane was processed for immunoreaction using mouse anti-His antibody (GE Health Care) diluted 1:10,000. Immunoreactive proteins were detected using a chemiluminescence kit (Biological Industries) according to the manufacturer’s protocol.
Growth curves
hipB+ cells (MGNR) carrying pZA21HipA plasmid and either an empty pQE30 vector or a pGltX plasmid were grown in LB medium at 37 °C with shaking. The optical density was measured in a multiplate reader (Victor2, Perkin-Elmer) every 2.5 min. At OD630 of 0.01, aTc was added at indicated concentrations to induce the expression of HipA. After 3 h, cells were diluted and plated on LB Petri dishes containing Kanamycin and Ampicillin and colony formation was monitored at 32 °C using the ScanLag setup22.
Death curves
hipB+ cells (MGNR) carrying pZA21HipA plasmid and either an empty pQE30 vector or a pGltX plasmid were grown in LB medium at 3 °C with shaking. At OD600 of 0.2, aTc (0 ng ml−1 or 4 ng ml−1) was added to induce the expression of HipA. After 3 h, cells were diluted into fresh medium containing Norfloxacin (8 μg ml−1). Cells were diluted on LB plates (without Norfloxacin) 0, 2.5, 5 and 24 h after the dilution into Norfloxacin. Colony forming units were counted after 12, 26 and 56 h of incubation at 37 °C. Until the addition of Norfloxacin, all strains, with and without the addition of aTc, showed equal growth curves, unless indicated otherwise. Statistical analysis was performed by the calculation of the percent of the survival cells after 24 h on Norfloxacin from four different death curves.
MS measurements and protein identification
His-GltX was purified as described in ‘Protein labelling, purification and analysis’, and separated by 12% SDS-polyacrylamide gel electrophoresis. We have purified GltX from cells overexpressing HipA and from cells that did not express HipA at all. The MS/MS study was done on these two protein samples. The proteins were reduced (3 mM dithiothreitol), modified with 12 mM iodoacetamide and digested in-gel with trypsin and chymotrypsin (Promega) separately at a 1:50 enzyme-to-substrate ratio in 10 mm ammonium bicarbonate and 10% acetonitrile.
The resulting peptides were resolved by reverse-phase chromatography on 0.075 × 200-mm fused silica capillaries (J&W) packed with Reprosil reversed-phase material (Dr Maisch GmbH, Germany). The peptides were eluted with 65 min linear gradients of 5–45% and 15 min at 95% acetonitrile with 0.1% formic acid in water at flow rates of 0.25 μl min−1. Mass spectrometry was performed by an ion-trap mass spectrometer (Orbitrap XL, Thermo) in a positive mode using repetitively full MS scan followed by collision-induced dissociation of the seven most dominant ions selected from the first MS scan. In order to analyze phosphopeptides, multistage activation was employed.
The mass spectrometry data were analyzed using the Proteome discoverer 1.3 (ThermoFisher Inc.) using both Sequest and Mascot search engines, searching against the E. coli section of the NCBI_NR database and versus the specific sequence of the protein. Identifications were filtered according to mass accuracy and 1% false discovery rate.
Northern blot analysis
ΔhipA hipB+ cells (MGNR) cells carrying pZA21HipA-mcherry plasmid13 and pGltX plasmid, as indicated in the text, were grown in LB medium with the appropriate antibiotics. At OD600 0.3, 1.2 μg ml−1 of IPTG was added when necessary for the induction of GltX and incubated for 30 min, after which HipA was induced by the addition of 20 ng ml−1 of aTc for 40 min. RNA was purified using the RNeasy kit (Qiagen) at pH 5.5. The samples were fractioned on a 18% polyacrylamide gel (19:1 acrylamide:bisacrylamide) containing 8 M urea. The samples were then transferred onto a Hybond+ membrane (GE Healthcare, Germany). tRNAGlu probe % 5′-(GCGGTGTCCTGGGCCTCT)-3′ was labeled and detected using ECL DIRECT Nucleic Labeling and Detection System (GE Healthcare, Germany).
Cloning GltX into pSA11
The GltX gene was cloned into EcoRI/XbaI site of pSA11 vector. GltX was amplified by GltX-FW: 5′-TCTCTCTCGAATTCAGGAGGAGCTCATCATGAAAATCAAAACTCGCTTCG-3′ GltX-Rev: 5′-TCTCTCTCTCTAGACCGTCTCGATATTGACGAATCA-3′.
As template, we used the plasmids purified from the library screen.
Additional information
How to cite this article: Kaspy, I. et al. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 4:3001 doi: 10.1038/ncomms4001 (2013).
References
Balaban, N. Q. Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev. 21, 768–775 (2011).
Lewis, K. Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010).
Bigger, J. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244, 497–500 (1944).
Keren, I., Shah, D., Spoering, A., Kaldalu, N. & Lewis, K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180 (2004).
Pearl, S., Gabay, C., Kishony, R., Oppenheim, A. & Balaban, N. Q. Nongenetic individuality in the host-phage interaction. PLoS Biol. 6, e120 (2008).
Scherrer, R. & Moyed, H. S. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 170, 3321–3326 (1988).
Maisonneuve, E., Shakespeare, L. J., Jorgensen, M. G. & Gerdes, K. Bacterial persistence by RNA endonucleases. Proc. Natl Acad. Sci. USA 108, 13206–13211 (2011).
Hayes, F. & Van Melderen, L. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 46, 386–408 (2011).
Moyed, H. S. & Bertrand, K. P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155, 768–775 (1983).
Correia, F. F. et al. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 188, 8360–8367 (2006).
Schumacher, M. A. et al. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323, 396–401 (2009).
Korch, S. B. & Hill, T. M. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188, 3826–3836 (2006).
Rotem, E. et al. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl Acad. Sci. USA 107, 12541–12546 (2010).
Korch, S. B., Henderson, T. A. & Hill, T. M. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50, 1199–1213 (2003).
Nguyen, D. et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986 (2011).
Amato, S. M., Orman, M. A. & Brynildsen, M. P. Metabolic control of persister formation in Escherichia coli. Mol. Cell 50, 475–487 (2013).
Maisonneuve, E., Castro-Camargo, M. & Gerdes, K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150 (2013).
Bokinsky, G. et al. HipA-triggered growth arrest and beta-lactam tolerance in Escherichia coli is mediated by RelA-dependent ppGpp synthesis. J. Bacteriol. 195, 3173–3182 (2013).
Young, F. S. & Neidhardt, F. C. Effect of inhibitors of elongation factor Tu on the metabolic regulation of protein synthesis in Escherichia coli. J. Bacteriol. 135, 675–686 (1978).
Ulbrandt, N. D., Newitt, J. A. & Bernstein, H. D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88, 187–196 (1997).
Cashel, M. & Gallant, J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841 (1969).
Levin-Reisman, I. et al. Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nat. Methods 7, 737–739 (2010).
Tosa, T. & Pizer, L. I. Biochemical bases for the antimetabolite action of L-serine hydroxamate. J. Bacteriol. 106, 972–982 (1971).
Sekine, S. et al. ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. EMBO J. 22, 676–688 (2003).
Germain, E., Castro-Roa, D., Zenkin, N. & Gerdes, K. Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254 (2013).
Potrykus, K. & Cashel, M. p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51 (2008).
Gerdes, K. & Maisonneuve, E. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 66, 103–123 (2012).
Aizenman, E., Engelberg-Kulka, H. & Glaser, G. An Escherichia coli chromosomal ‘addiction module’ regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl Acad. Sci. USA 93, 6059–6063 (1996).
Kuroda, A., Murphy, H., Cashel, M. & Kornberg, A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272, 21240–21243 (1997).
Kuroda, A. et al. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293, 705–708 (2001).
Hansen, S. et al. Regulation of the Escherichia coli HipBA toxin-antitoxin system by proteolysis. PLoS One 7, e39185 (2012).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Wexselblatt, E. et al. Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog. 8, e1002925 (2012).
Allison, K. R., Brynildsen, M. P. & Collins, J. J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220 (2011).
Luidalepp, H., Joers, A., Kaldalu, N. & Tenson, T. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J. Bacteriol. 193, 3598–3605 (2011).
Hansen, S., Lewis, K. & Vulic, M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother 52, 2718–2726 (2008).
Fasani, R. A. & Savageau, M. A. Molecular mechanisms of multiple toxin-antitoxin systems are coordinated to govern the persister phenotype. Proc. Natl Acad. Sci. USA 110, E2528–E2537 (2013).
Gefen, O. & Balaban, N. Q. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS. Microbiol. Rev. 33, 704–717 (2009).
Dahl, J. L. et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl Acad. Sci. USA 100, 10026–10031 (2003).
Neidhardt, F. C., Bloch, P. L. & Smith, D. F. Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 (1974).
Acknowledgements
We thank Hila Wolf-Levi from the Selmor Proteomics Center in the Haifa University, for the MS-MS analysis; Ofra Moshel and Alex Eliassaf Feldstein for helping the MS-MS interpretations. Yoav Smith for statistical assistance. NQB is supported by the European Research Council grant no. 260871. G.G. and N.Q.B. acknowledge funding of the Israel Science Foundation grant no. 374/08 and no. 592/10, respectively.
Author information
Authors and Affiliations
Contributions
N.Q.B and G.G. designed the project. I.K. performed most of the experimental work. E.R. performed the survival and persisters assays. N.W. derived the model and equations. I.R. was responsible for the cloning of the different genes. I.K., N.Q.B and G.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1-S5, Supplementary Tables S1-S3 and Supplementary References (PDF 1049 kb)
Rights and permissions
About this article
Cite this article
Kaspy, I., Rotem, E., Weiss, N. et al. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun 4, 3001 (2013). https://doi.org/10.1038/ncomms4001
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms4001
This article is cited by
-
Glyphosate affects persistence and tolerance but not antibiotic resistance
BMC Microbiology (2023)
-
Shaping bacterial gene expression by physiological and proteome allocation constraints
Nature Reviews Microbiology (2023)
-
Molecular mechanism of toxin neutralization in the HipBST toxin-antitoxin system of Legionella pneumophila
Nature Communications (2022)
-
Biology and evolution of bacterial toxin–antitoxin systems
Nature Reviews Microbiology (2022)
-
Mutations in respiratory complex I promote antibiotic persistence through alterations in intracellular acidity and protein synthesis
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.