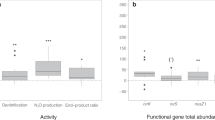
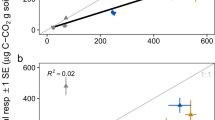
Abstract
Nitrous oxide (N2O) is the predominant ozone-depleting substance and contributes approximately 6% to overall global warming1,2. Terrestrial ecosystems account for nearly 70% of total global N2O atmospheric loading, of which at least 45% can be attributed to microbial cycling of nitrogen in agriculture3. The reduction of N2O to nitrogen gas by microorganisms is critical for mitigating its emissions from terrestrial ecosystems, yet the determinants of a soil’s capacity to act as a source or sink for N2O remain uncertain4. Here, we demonstrate that the soil N2O sink capacity is mostly explained by the abundance and phylogenetic diversity of a newly described N2O-reducing microbial group5,6, which mediate the influence of edaphic factors. Analyses of interactions and niche preference similarities suggest niche differentiation or even competitive interactions between organisms with the two types of N2O reductase. We further identified several recurring communities comprised of co-occurring N2O-reducing bacterial genotypes that were significant indicators of the soil N2O sink capacity across different European soils.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ravishankara, A. R., Daniel, J. S. & Portmann, R. W. Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125 (2009).
Montzka, S. A., Dlugokencky, E. J. & Butler, J. H. Non-CO2 greenhouse gases and climate change. Nature 476, 43–50 (2011).
Syakila, A. & Kroeze, C. The global nitrous oxide budget revisited. Greenhouse Gas Meas. Manage. 1, 17–26 (2011).
Butterbach-Bahl, K., Baggs, E. M., Dannenmann, M., Kiese, R. & Zechmeister-Boltenstern, S. Nitrous oxide emissions from soils: How well do we understand the processes and their controls? Phil. Trans. R. Soc. B. 368, 20130122 (2013).
Jones, C. M., Graf, D. R. H., Bru, D., Philippot, L. & Hallin, S. The unaccounted yet abundant nitrous oxide-reducing microbial community: A potential nitrous oxide sink. ISME J. 7, 417–426 (2013).
Sanford, R. A. et al. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl Acad. Sci. USA 109, 19709–19714 (2012).
Hofmann, D. J. et al. The role of carbon dioxide in climate forcing from 1979 to 2004: Introduction of the annual greenhouse gas index. Tellus B 58, 614–619 (2006).
Chapuis-Lardy, L., Wrage, N., Metay, A., Chotte, J. & Bernoux, M. Soils, a sink for N2O? A review. Glob. Change Biol. 13, 1–17 (2007).
Richardson, D., Felgate, H., Watmough, N., Thomson, A. & Baggs, E. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle—could enzymic regulation hold the key? Trends Biotechnol. 27, 388–397 (2009).
Spiro, S. Nitrous oxide production and consumption: Regulation of gene expression by gas-sensitive transcription factors. Phil. Trans. R. Soc. B 367, 1213–1225 (2012).
Jones, C. M., Stres, B., Rosenquist, M. & Hallin, S. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25, 1955–1966 (2008).
Zumft, W. & Kroneck, P. Respiratory transformation of nitrous oxide (N2O) to dinitrogen by Bacteria and Archaea. Adv. Microb. Physiol. 52, 107–227 (2007).
Philippot, L., Andert, J., Jones, C. M., Bru, D. & Hallin, S. Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Glob. Change Biol. 17, 1497–1504 (2011).
Wood, D. et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294, 2317–2323 (2001).
Petersen, D. G. et al. Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ. Microbiol. 14, 993–1008 (2012).
Jones, C. M. & Hallin, S. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 4, 633–641 (2010).
Enwall, K., Throbäck, I. N., Stenberg, M., Söderström, M. & Hallin, S. Soil resources influence spatial patterns of denitrifying communities at scales compatible with land management. Appl. Environ. Microbiol. 76, 2243–2250 (2010).
Philippot, L. et al. Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ. Microbiol. 11, 1518–1526 (2009).
Braker, G., Dorsch, P. & Bakken, L. R. Genetic characterization of denitrifier communities with contrasting intrinsic functional traits. FEMS Microbiol. Ecol. 79, 542–554 (2012).
Philippot, L. et al. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 7, 1609–1619 (2013).
Cuhel, J. & Simek, M. Proximal and distal control by pH of denitrification rate in a pasture soil. Agr. Ecosyst. Environ. 141, 230–233 (2011).
Baggs, E. M., Smales, C. L. & Bateman, E. J. Changing pH shifts the microbial source as well as the magnitude of N2O emission from soil. Biol. Fert. Soils 46, 793–805 (2010).
Firestone, M. K., Firestone, R. B. & Tiedje, J. M. Nitrous oxide from soil denitrification: Factors controlling its biological production. Science 208, 749–751 (1980).
Penton, C. R. et al. Functional genes to assess nitrogen cycling and aromatic hydrocarbon degradation: Primers and processing matter. Front. Microbiol. 4, 279 (2013).
Matsen, F. A., Kodner, R. B. & Armbrust, E. V. pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11, 538 (2010).
Faith, D. P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10 (1992).
Rosseel, Y. lavaan: An R package for structural equation modeling. J. Stat. Softw. 48, 1–36 (2012).
Grace, J. B. & Bollen, K. A. Representing general theoretical concepts in structural equation models: The role of composite variables. Environ. Ecol. Stat. 15, 191–213 (2007).
Stone, E. A. & Ayroles, J. F. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 5, e1000479 (2009).
Strobl, C., Boulesteix, A. L., Kneib, T. & Augustin, T. Conditional variable importance for random forests. BMC Bioinformatics 9, 307 (2008).
Acknowledgements
We thank all of our collaborators who contributed to the soil sampling. This research was supported by the European Commission within the EcoFINDERS project (FP7-264465), the French Embassy in Dublin, the Conseil Régional de Bourgogne, Teagasc and The Swedish Research Council Formas (2009-741).
Author information
Authors and Affiliations
Contributions
C.M.J., A.S., S.H. and L.P. designed the study, analysed the data and compiled the manuscript with the help of B.G. and P.L. Soil samples were collected by D.B., F.P.B., C.M.J., A.S. and L.P. with support from the EcoFINDERS project. Microcosm set-up and gas analysis was performed by M-C.B., D.B., F.P.B. and C.M.J., and soil DNA extractions, real-time PCR and 454 sequencing was performed by A.S., M-C.B. and D.B.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Jones, C., Spor, A., Brennan, F. et al. Recently identified microbial guild mediates soil N2O sink capacity. Nature Clim Change 4, 801–805 (2014). https://doi.org/10.1038/nclimate2301
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nclimate2301
This article is cited by
-
How to adequately represent biological processes in modeling multifunctionality of arable soils
Biology and Fertility of Soils (2024)
-
Soil microbiomes and one health
Nature Reviews Microbiology (2023)
-
Denitrifying Woodchip Bioreactors: A Microbial Solution for Nitrate in Agricultural Wastewater—A Review
Journal of Microbiology (2023)
-
Niche Differentiation Among Canonical Nitrifiers and N2O Reducers Is Linked to Varying Effects of Nitrification Inhibitors DCD and DMPP in Two Arable Soils
Microbial Ecology (2023)
-
Soil water extract and bacteriome determine N2O emission potential in soils
Biology and Fertility of Soils (2023)