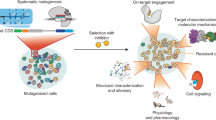
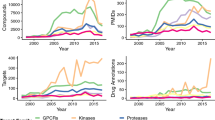
Abstract
Target-identification and mechanism-of-action studies have important roles in small-molecule probe and drug discovery. Biological and technological advances have resulted in the increasing use of cell-based assays to discover new biologically active small molecules. Such studies allow small-molecule action to be tested in a more disease-relevant setting at the outset, but they require follow-up studies to determine the precise protein target or targets responsible for the observed phenotype. Target identification can be approached by direct biochemical methods, genetic interactions or computational inference. In many cases, however, combinations of approaches may be required to fully characterize on-target and off-target effects and to understand mechanisms of small-molecule action.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sundberg, S.A. High-throughput and ultra-high-throughput screening: solution- and cell-based approaches. Curr. Opin. Biotechnol. 11, 47–53 (2000).
Mayr, L.M. & Bojanic, D. Novel trends in high-throughput screening. Curr. Opin. Pharmacol. 9, 580–588 (2009).
Koehn, F.E. High impact technologies for natural products screening. Prog. Drug Res. 65, 175, 177–210 (2008).
Lachance, H., Wetzel, S., Kumar, K. & Waldmann, H. Charting, navigating, and populating natural product chemical space for drug discovery. J. Med. Chem. 55, 5989–6001 (2012).
Nielsen, T.E. & Schreiber, S.L. Towards the optimal screening collection: a synthesis strategy. Angew. Chem. Int. Edn Engl. 47, 48–56 (2008).
O'Connor, C.J., Beckmann, H.S. & Spring, D.R. Diversity-oriented synthesis: producing chemical tools for dissecting biology. Chem. Soc. Rev. 41, 4444–4456 (2012).
Swinney, D.C. & Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 10, 507–519 (2011).
Terstappen, G.C., Schlupen, C., Raggiaschi, R. & Gaviraghi, G. Target deconvolution strategies in drug discovery. Nat. Rev. Drug Discov. 6, 891–903 (2007).
García-García, M.J. et al. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc. Natl. Acad. Sci. USA 102, 5913–5919 (2005).
Muto, A. et al. Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 1, e66 (2005).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Pickart, M.A. et al. Genome-wide reverse genetics framework to identify novel functions of the vertebrate secretome. PLoS ONE 1, e104 (2006).
Ecker, A., Bushell, E.S., Tewari, R. & Sinden, R.E. Reverse genetics screen identifies six proteins important for malaria development in the mosquito. Mol. Microbiol. 70, 209–220 (2008).
Mitchison, T.J. Towards a pharmacological genetics. Chem. Biol. 1, 3–6 (1994).
Schreiber, S.L. Chemical genetics resulting from a passion for synthetic organic chemistry. Bioorg. Med. Chem. 6, 1127–1152 (1998).
Inglese, J. et al. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 3, 466–479 (2007).
Macarron, R. et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 10, 188–195 (2011).
Wyatt, P.G., Gilbert, I.H., Read, K.D. & Fairlamb, A.H. Target validation: linking target and chemical properties to desired product profile. Curr. Top. Med. Chem. 11, 1275–1283 (2011).
Kauselmann, G., Dopazo, A. & Link, W. Identification of disease-relevant genes for molecularly-targeted drug discovery. Curr. Cancer Drug Targets 12, 1–13 (2012).
Stockwell, B.R. Chemical genetics: ligand-based discovery of gene function. Nat. Rev. Genet. 1, 116–125 (2000).
Stockwell, B.R. Exploring biology with small organic molecules. Nature 432, 846–854 (2004).
Clemons, P.A. Complex phenotypic assays in high-throughput screening. Curr. Opin. Chem. Biol. 8, 334–338 (2004).
Schreiber, S.L. & Crabtree, G.R. The mechanism of action of cyclosporin A and FK506. Immunol. Today 13, 136–142 (1992).
Harding, M.W., Galat, A., Uehling, D.E. & Schreiber, S.L. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 341, 758–760 (1989).
Liu, J. et al. Inhibition of T cell signaling by immunophilin-ligand complexes correlates with loss of calcineurin phosphatase activity. Biochemistry 31, 3896–3901 (1992).
Brown, E.J. et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369, 756–758 (1994).
Yoshida, M., Nomura, S. & Beppu, T. Effects of trichostatins on differentiation of murine erythroleukemia cells. Cancer Res. 47, 3688–3691 (1987).
Yoshida, M., Kijima, M., Akita, M. & Beppu, T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17174–17179 (1990).
Taunton, J., Hassig, C.A. & Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 (1996).
McNamara, C. & Winzeler, E.A. Target identification and validation of novel antimalarials. Future Microbiol. 6, 693–704 (2011).
Whitebread, S., Hamon, J., Bojanic, D. & Urban, L. Keynote review: in vitro safety pharmacology profiling: an essential tool for successful drug development. Drug Discov. Today 10, 1421–1433 (2005).
Xie, L. & Bourne, P.E. Structure-based systems biology for analyzing off-target binding. Curr. Opin. Struct. Biol. 21, 189–199 (2011).
Chen, S. et al. Self-renewal of embryonic stem cells by a small molecule. Proc. Natl. Acad. Sci. USA 103, 17266–17271 (2006).
Apsel, B. et al. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat. Chem. Biol. 4, 691–699 (2008).
Ito, T. et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010).
Knight, Z.A., Lin, H. & Shokat, K.M. Targeting the cancer kinome through polypharmacology. Nat. Rev. Cancer 10, 130–137 (2010).
Burdine, L. & Kodadek, T. Target identification in chemical genetics: the (often) missing link. Chem. Biol. 11, 593–597 (2004).
Zheng, X.S., Chan, T.F. & Zhou, H.H. Genetic and genomic approaches to identify and study the targets of bioactive small molecules. Chem. Biol. 11, 609–618 (2004).
Weinstein, J.N. et al. An information-intensive approach to the molecular pharmacology of cancer. Science 275, 343–349 (1997).
Hughes, T.R. et al. Functional discovery via a compendium of expression profiles. Cell 102, 109–126 (2000).
Young, D.W. et al. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nat. Chem. Biol. 4, 59–68 (2008).
Fomina-Yadlin, D. et al. Small-molecule inducers of insulin expression in pancreatic alpha-cells. Proc. Natl. Acad. Sci. USA 107, 15099–15104 (2010).
Cuatrecasas, P., Wilchek, M. & Anfinsen, C.B. Selective enzyme purification by affinity chromatography. Proc. Natl. Acad. Sci. USA 61, 636–643 (1968).
Hirota, T. et al. Identification of small molecule activators of cryptochrome. Science 337, 1094–1097 (2012).
Brehmer, D., Godl, K., Zech, B., Wissing, J. & Daub, H. Proteome-wide identification of cellular targets affected by bisindolylmaleimide-type protein kinase C inhibitors. Mol. Cell Proteomics 3, 490–500 (2004).
Wissing, J. et al. Chemical proteomic analysis reveals alternative modes of action for pyrido[2,3-d]pyrimidine kinase inhibitors. Mol. Cell Proteomics 3, 1181–1193 (2004).
Oda, Y. et al. Quantitative chemical proteomics for identifying candidate drug targets. Anal. Chem. 75, 2159–2165 (2003).
Wang, G., Shang, L., Burgett, A.W., Harran, P.G. & Wang, X. Diazonamide toxins reveal an unexpected function for ornithine delta-amino transferase in mitotic cell division. Proc. Natl. Acad. Sci. USA 104, 2068–2073 (2007).
Ong, S.E. et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc. Natl. Acad. Sci. USA 106, 4617–4622 (2009).
Fleischer, T.C. et al. Chemical proteomics identifies Nampt as the target of CB30865, an orphan cytotoxic compound. Chem. Biol. 17, 659–664 (2010).
Shiyama, T., Furuya, M., Yamazaki, A., Terada, T. & Tanaka, A. Design and synthesis of novel hydrophilic spacers for the reduction of nonspecific binding proteins on affinity resins. Bioorg. Med. Chem. 12, 2831–2841 (2004).
Speers, A.E. & Cravatt, B.F. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J. Am. Chem. Soc. 127, 10018–10019 (2005).
van der Veken, P. et al. Development of a novel chemical probe for the selective enrichment of phosphorylated serine- and threonine-containing peptides. Chembiochem. 6, 2271–2280 (2005).
Fonoviisć, M., Verhelst, S.H., Sorum, M.T. & Bogyo, M. Proteomics evaluation of chemically cleavable activity-based probes. Mol. Cell Proteomics 6, 1761–1770 (2007).
Verhelst, S.H., Fonovic, M. & Bogyo, M. A mild chemically cleavable linker system for functional proteomic applications. Angew. Chem. Int. Ed. Engl. 46, 1284–1286 (2007).
Evans, M.J., Saghatelian, A., Sorensen, E.J. & Cravatt, B.F. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat. Biotechnol. 23, 1303–1307 (2005).
Cisar, J.S. & Cravatt, B.F. Fully functionalized small-molecule probes for integrated phenotypic screening and target identification. J. Am. Chem. Soc. 134, 10385–10388 (2012).
Park, J., Oh, S. & Park, S.B. Discovery and target identification of an antiproliferative agent in live cells using fluorescence difference in two-dimensional gel electrophoresis. Angew. Chem. Int. Ed. Engl. 51, 5447–5451 (2012).
Kawatani, M. et al. The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I. Proc. Natl. Acad. Sci. USA 105, 11691–11696 (2008).
Saxena, C. et al. Capture of drug targets from live cells using a multipurpose immuno-chemo-proteomics tool. J. Proteome Res. 8, 3951–3957 (2009).
Khersonsky, S.M. et al. Facilitated forward chemical genetics using a tagged triazine library and zebrafish embryo screening. J. Am. Chem. Soc. 125, 11804–11805 (2003).
Kim, Y.K. & Chang, Y.T. Tagged library approach facilitates forward chemical genetics. Mol. Biosyst. 3, 392–397 (2007).
Tao, S.C., Chen, C.S. & Zhu, H. Applications of protein microarray technology. Comb. Chem. High Throughput Screen. 10, 706–718 (2007).
Lomenick, B., Olsen, R.W. & Huang, J. Identification of direct protein targets of small molecules. ACS Chem. Biol. 6, 34–46 (2011).
Chan, J.N. et al. Target identification by chromatographic co-elution: monitoring of drug-protein interactions without immobilization or chemical derivatization. Mol. Cell. Proteomics 11, M111.016642 (2012).
Aebersold, R. & Mann, M. Mass spectrometry–based proteomics. Nature 422, 198–207 (2003).
Ong, S.E. & Mann, M. Mass spectrometry–based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 (2005).
Blagoev, B. et al. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 (2003).
Ranish, J.A. et al. The study of macromolecular complexes by quantitative proteomics. Nat. Genet. 33, 349–355 (2003).
Rix, U. & Superti-Furga, G. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 5, 616–624 (2009).
Ong, S.E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 (2002).
Daub, H. et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 (2008).
Bendall, S.C. et al. Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol. Cell Proteomics 7, 1587–1597 (2008).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 (1999).
Ross, P.L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169 (2004).
Bantscheff, M. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 (2007).
Thompson, A. et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 (2003); erratum 75, 4942 (2003); erratum 78, 4235 (2006).
Hsu, J.L., Huang, S.Y., Chow, N.H. & Chen, S.H. Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 75, 6843–6852 (2003).
Boersema, P.J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A.J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Mertins, P. et al. iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics. Mol. Cell Proteomics 11, M111.014423 (2012).
Mortensen, P. et al. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J. Proteome Res. 9, 393–403 (2010).
Tsou, C.C. et al. MaXIC-Q Web: a fully automated web service using statistical and computational methods for protein quantitation based on stable isotope labeling and LC-MS. Nucleic Acids Res. 37, W661–W669 (2009).
Cox, J. et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Margolin, A.A. et al. Empirical Bayes analysis of quantitative proteomics experiments. PLoS ONE 4, e7454 (2009).
Fabian, M.A. et al. A small molecule–kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 23, 329–336 (2005).
Boutros, M. & Ahringer, J. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 9, 554–566 (2008).
Perlstein, E.O., Ruderfer, D.M., Roberts, D.C., Schreiber, S.L. & Kruglyak, L. Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat. Genet. 39, 496–502 (2007).
Pierce, S.E. et al. A unique and universal molecular barcode array. Nat. Methods 3, 601–603 (2006).
Roemer, T. & Boone, C. Systems-level antimicrobial drug and drug synergy discovery. Nat. Chem. Biol. 9, 222–231 (2013).
Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006).
Wang, J. et al. Cellular phenotype recognition for high-content RNA interference genome-wide screening. J. Biomol. Screen. 13, 29–39 (2008).
Eggert, U.S. et al. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2, e379 (2004).
Guertin, D.A., Guntur, K.V., Bell, G.W., Thoreen, C.C. & Sabatini, D.M. Functional genomics identifies TOR-regulated genes that control growth and division. Curr. Biol. 16, 958–970 (2006).
Knight, Z.A. & Shokat, K.M. Chemical genetics: where genetics and pharmacology meet. Cell 128, 425–430 (2007).
Castoreno, A.B. et al. Small molecules discovered in a pathway screen target the Rho pathway in cytokinesis. Nat. Chem. Biol. 6, 457–463 (2010).
Wacker, S.A., Houghtaling, B.R., Elemento, O. & Kapoor, T.M. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat. Chem. Biol. 8, 235–237 (2012).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Iorio, F. et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci. USA 107, 14621–14626 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Feng, Y., Mitchison, T.J., Bender, A., Young, D.W. & Tallarico, J.A. Multi-parameter phenotypic profiling: using cellular effects to characterize small-molecule compounds. Nat. Rev. Drug Discov. 8, 567–578 (2009).
Wagner, B.K. & Clemons, P.A. Connecting synthetic chemistry decisions to cell and genome biology using small-molecule phenotypic profiling. Curr. Opin. Chem. Biol. 13, 539–548 (2009).
Haupt, V.J. & Schroeder, M. Old friends in new guise: repositioning of known drugs with structural bioinformatics. Brief. Bioinform. 12, 312–326 (2011).
Koutsoukas, A. et al. From in silico target prediction to multi-target drug design: current databases, methods and applications. J. Proteomics 74, 2554–2574 (2011).
Barrett, T. et al. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 35, D760–D765 (2007).
Seiler, K.P. et al. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 36, D351–D359 (2008).
Wang, Y. et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 37, W623–W633 (2009).
Stegmaier, K. et al. Gene expression–based high-throughput screening (GE-HTS) and application to leukemia differentiation. Nat. Genet. 36, 257–263 (2004).
Hieronymus, H. et al. Gene expression signature–based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 10, 321–330 (2006).
Kim, Y.K. et al. Relationship of stereochemical and skeletal diversity of small molecules to cellular measurement space. J. Am. Chem. Soc. 126, 14740–14745 (2004).
Paull, K.D. et al. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 81, 1088–1092 (1989).
Zaharevitz, D.W. et al. Discovery and initial characterization of the paullones, a novel class of small-molecule inhibitors of cyclin-dependent kinases. Cancer Res. 59, 2566–2569 (1999).
Marton, M.J. et al. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat. Med. 4, 1293–1301 (1998).
Ganter, B. et al. Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. J. Biotechnol. 119, 219–244 (2005).
Fliri, A.F., Loging, W.T., Thadeio, P.F. & Volkmann, R.A. Biological spectra analysis: Linking biological activity profiles to molecular structure. Proc. Natl. Acad. Sci. USA 102, 261–266 (2005).
Berg, E.L., Kunkel, E.J., Hytopoulos, E. & Plavec, I. Characterization of compound mechanisms and secondary activities by BioMAP analysis. J. Pharmacol. Toxicol. Methods 53, 67–74 (2006).
Kauvar, L.M. et al. Predicting ligand binding to proteins by affinity fingerprinting. Chem. Biol. 2, 107–118 (1995).
Campillos, M., Kuhn, M., Gavin, A.C., Jensen, L.J. & Bork, P. Drug target identification using side-effect similarity. Science 321, 263–266 (2008).
Dixon, S.L. & Villar, H.O. Bioactive diversity and screening library selection via affinity fingerprinting. J. Chem. Inf. Comput. Sci. 38, 1192–1203 (1998).
Perlman, Z.E. et al. Multidimensional drug profiling by automated microscopy. Science 306, 1194–1198 (2004).
Carpenter, A.E. Image-based chemical screening. Nat. Chem. Biol. 3, 461–465 (2007).
Chen, B., Wild, D. & Guha, R. PubChem as a source of polypharmacology. J. Chem. Inf. Model. 49, 2044–2055 (2009).
Tanikawa, T. et al. Using biological performance similarity to inform disaccharide library design. J. Am. Chem. Soc. 131, 5075–5083 (2009).
Plouffe, D. et al. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc. Natl. Acad. Sci. USA 105, 9059–9064 (2008).
Wolpaw, A.J. et al. Modulatory profiling identifies mechanisms of small molecule-induced cell death. Proc. Natl. Acad. Sci. USA 108, E771–E780 (2011).
Cheng, T., Li, Q., Wang, Y. & Bryant, S.H. Identifying compound-target associations by combining bioactivity profile similarity search and public databases mining. J. Chem. Inf. Model. 51, 2440–2448 (2011).
Petrone, P.M. et al. Rethinking molecular similarity: comparing compounds on the basis of biological activity. ACS Chem. Biol. 7, 1399–1409 (2012).
Nidhi, Glick, M, Davies, J.W. & Jenkins, J.L. Prediction of biological targets for compounds using multiple-category Bayesian models trained on chemogenomics databases. J. Chem. Inf. Model. 46, 1124–1133 (2006).
Paolini, G.V., Shapland, R.H., van Hoorn, W.P., Mason, J.S. & Hopkins, A.L. Global mapping of pharmacological space. Nat. Biotechnol. 24, 805–815 (2006).
Keiser, M.J. et al. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 25, 197–206 (2007).
Lounkine, E. et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature 486, 361–367 (2012).
Keiser, M.J. et al. Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009).
Gregori-Puigjané, E. et al. Identifying mechanism-of-action targets for drugs and probes. Proc. Natl. Acad. Sci. USA 109, 11178–11183 (2012).
Laggner, C. et al. Chemical informatics and target identification in a zebrafish phenotypic screen. Nat. Chem. Biol. 8, 144–146 (2012).
Oprea, T.I., Tropsha, A., Faulon, J.L. & Rintoul, M.D. Systems chemical biology. Nat. Chem. Biol. 3, 447–450 (2007).
Hopkins, A.L. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 4, 682–690 (2008).
Boran, A.D. & Iyengar, R. Systems pharmacology. Mt. Sinai J. Med. 77, 333–344 (2010).
Yamanishi, Y., Araki, M., Gutteridge, A., Honda, W. & Kanehisa, M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 24, i232–i240 (2008).
He, Z. et al. Predicting drug-target interaction networks based on functional groups and biological features. PLoS ONE 5, e9603 (2010).
Mei, J.P., Kwoh, C.K., Yang, P., Li, X.L. & Zheng, J. Drug-target interaction prediction by learning from local information and neighbors. Bioinformatics 29, 238–245 (2013).
Bach, S. et al. Roscovitine targets, protein kinases and pyridoxal kinase. J. Biol. Chem. 280, 31208–31219 (2005).
Kuai, L. et al. AAK1 identified as an inhibitor of neuregulin-1/ErbB4–dependent neurotrophic factor signaling using integrative chemical genomics and proteomics. Chem. Biol. 18, 891–906 (2011).
Raj, L. et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature 475, 231–234 (2011).
Arastu-Kapur, S. et al. Nonproteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: a link to clinical adverse events. Clin. Cancer Res. 17, 2734–2743 (2011).
Wen, Q. et al. Identification of regulators of polyploidization presents therapeutic targets for treatment of AMKL. Cell 150, 575–589 (2012).
Winter, G.E. et al. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nat. Chem. Biol. 8, 905–912 (2012).
Parsons, A.B. et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22, 62–69 (2004).
Perlstein, E.O. et al. Revealing complex traits with small molecules and naturally recombinant yeast strains. Chem. Biol. 13, 319–327 (2006).
Acknowledgements
This work was supported by US National Institutes of Health Genomics Based Drug Discovery–Target ID Project grant RL1HG004671, which is administratively linked to the US National Institutes of Health grants RL1CA133834, RL1GM084437 and UL1RR024924.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Schenone, M., Dančík, V., Wagner, B. et al. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol 9, 232–240 (2013). https://doi.org/10.1038/nchembio.1199
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1199
This article is cited by
-
Kallistatin deficiency exacerbates neuronal damage after cardiac arrest
Scientific Reports (2024)
-
One substrate many enzymes virtual screening uncovers missing genes of carnitine biosynthesis in human and mouse
Nature Communications (2024)
-
Synthesis and pharmacological evaluation of novel coumarin based triazolyl glycoconjugates as potential antibacterial and anti-proliferative agents
Medicinal Chemistry Research (2024)
-
Target identification of small molecules: an overview of the current applications in drug discovery
BMC Biotechnology (2023)
-
Targeting the PEDV 3CL protease for identification of small molecule inhibitors: an insight from virtual screening, ADMET prediction, molecular dynamics, free energy landscape, and binding energy calculations
Journal of Biological Engineering (2023)