Abstract
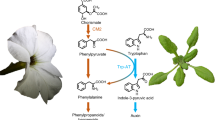
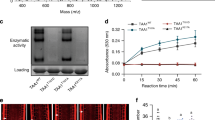
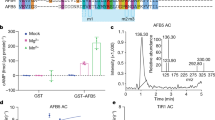
We identify an Arabidopsis pyridoxal-phosphate–dependent aminotransferase, VAS1, whose loss-of-function simultaneously increases amounts of the phytohormone auxin and the ethylene precursor 1-aminocyclopropane-1-carboxylate. VAS1 uses the ethylene biosynthetic intermediate methionine as an amino donor and the auxin biosynthetic intermediate indole-3-pyruvic acid as an amino acceptor to produce L-tryptophan and 2-oxo-4-methylthiobutyric acid. Our data indicate that VAS1 serves key roles in coordinating the amounts of these two vital hormones.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
References
Santner, A. & Estelle, M. Nature 459, 1071–1078 (2009).
Jaillais, Y. & Chory, J. Nat. Struct. Mol. Biol. 17, 642–645 (2010).
Franklin, K.A. New Phytol. 179, 930–944 (2008).
Li, L. et al. Genes Dev. 26, 785–790 (2012).
Tao, Y. et al. Cell 133, 164–176 (2008).
Stepanova, A.N. et al. Cell 133, 177–191 (2008).
Yamada, M., Greenham, K., Prigge, M.J., Jensen, P.J. & Estelle, M. Plant Physiol. 151, 168–179 (2009).
Stepanova, A.N. et al. Plant Cell 23, 3961–3973 (2011).
Won, C. et al. Proc. Natl. Acad. Sci. USA 108, 18518–18523 (2011).
Mashiguchi, K. et al. Proc. Natl. Acad. Sci. USA 108, 18512–18517 (2011).
Zhao, Y. Mol Plant 5, 334–338 (2012).
Liepman, A.H. & Olsen, L.I. Crit. Rev. Plant Sci. 23, 73–89 (2004).
Ulmasov, T., Murfett, J., Hagen, G. & Guilfoyle, T.J. Plant Cell 9, 1963–1971 (1997).
Albers, E. IUBMB Life 61, 1132–1142 (2009).
Bürstenbinder, K., Rzewuski, G., Wirtz, M., Hell, R. & Sauter, M. Plant J. 49, 238–249 (2007).
Baur, A.H. & Yang, S.F. Phytochemistry 11, 3207–3214 (1972).
Tsuchisaka, A. & Theologis, A. Plant Physiol. 136, 2982–3000 (2004).
Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S. & Ecker, J.R. Science 284, 2148–2152 (1999).
Pierik, R., Djakovic-Petrovic, T., Keuskamp, D.H., de Wit, M. & Voesenek, L.A. Plant Physiol. 149, 1701–1712 (2009).
Poeggeler, B. et al. Brain Res. 815, 382–388 (1999).
Chowdhury, G. et al. Chem. Res. Toxicol. 22, 1905–1912 (2009).
Lukowitz, W., Gillmor, C.S. & Scheible, W.R. Plant Physiol. 123, 795–805 (2000).
Nakagawa, T. et al. J. Biosci. Bioeng. 104, 34–41 (2007).
Jez, J.M., Ferrer, J.L., Bowman, M.E., Dixon, R.A. & Noel, J.P. Biochemistry 39, 890–902 (2000).
Smets, R., Claes, V., Van Onckelen, H.A. & Prinsen, E. J. Chromatogr. A 993, 79–87 (2003).
Edlund, A., Eklof, S., Sundberg, B., Moritz, T. & Sandberg, G. Plant Physiol. 108, 1043–1047 (1995).
Tam, Y.Y. & Normanly, J. J. Chromatogr. A 800, 101–108 (1998).
Kowalczyk, M. & Sandberg, G. Plant Physiol. 127, 1845–1853 (2001).
Rittenberg, D. & Foster, L. J. Biol. Chem. 133, 737–744 (1940).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. Mol. Biol. Evol. 24, 1596–1599 (2007).
Acknowledgements
We thank W. Chen for technical assistance and the Arabidopsis Biological Resource Center for mutant seeds. These studies were supported by US National Institutes of Health (NIH) grant 5R01GM52413 to J.C., the Internal Grant Agency of Palacký University (PrF_2012_016) and the Grant Agency of the Academy of Sciences of the Czech Republic (KAN200380801) to O.N., the Swedish Governmental Agency for Innovation Systems and the Swedish Research Council (K.L.), NIH grant R01GM68631 to Y.Z., the National Science Foundation under award nos. EEC-0813570 and MCB-0645794 to J.P.N. and the Howard Hughes Medical Institute (Z.Z., J.P.N., J.C.).
Author information
Authors and Affiliations
Contributions
Z.Z. and Y.G. designed research, performed research, analyzed data and wrote the paper. O.N. and K.L. measured the amount of IAA and 3-IPA and analyzed data. X.D. and Y.Z. provided yuc1-163 yuc4 seeds. J.P.N. and J.C. designed research, analyzed data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results (PDF 14253 kb)
Rights and permissions
About this article
Cite this article
Zheng, Z., Guo, Y., Novák, O. et al. Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol 9, 244–246 (2013). https://doi.org/10.1038/nchembio.1178
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1178
This article is cited by
-
Modulation of auxin formation by the cytosolic phenylalanine biosynthetic pathway
Nature Chemical Biology (2020)
-
Proteomic profiles during adventitious rooting of Eucalyptus species relevant to the cellulose industry
New Forests (2020)
-
Aromatic amino acid aminotransferases in plants
Phytochemistry Reviews (2018)
-
Regulation of seedling growth by ethylene and the ethylene–auxin crosstalk
Planta (2017)
-
The Arabidopsis phytohormone crosstalk network involves a consecutive metabolic route and circular control units of transcription factors that regulate enzyme-encoding genes
BMC Systems Biology (2016)