Abstract
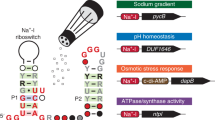
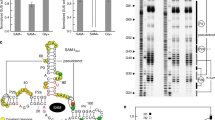
We report what is to our knowledge the first natural RNA that regulates gene expression in response to intracellular ATP. Using a biochemical screen, we found that several putative riboswitches bind ATP in vitro. The ydaO motif specifically bound ATP and regulated expression of endogenous and reporter genes in response to ATP concentrations in Bacillus subtilis. This discovery demonstrates a role for RNAs in regulating gene expression in response to energy balance in bacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Tarasov, A.I., Griffiths, E.J. & Rutter, G.A. Cell Calcium 52, 28–35 (2012).
Hardie, D.G., Ross, F.A. & Hawley, S.A. Nat. Rev. Mol. Cell Biol. 13, 251–262 (2012).
Sluse, F.E. Adv. Exp. Med. Biol. 942, 137–156 (2012).
Bastet, L., Dube, A., Masse, E. & Lafontaine, D.A. Mol. Microbiol. 80, 1148–1154 (2011).
Sudarsan, N. et al. Science 321, 411–413 (2008).
Sassanfar, M. & Szostak, J.W. Nature 364, 550–553 (1993).
Smith, A.M., Fuchs, R.T., Grundy, F.J. & Henkin, T.M. RNA Biol. 7, 104–110 (2010).
Weinberg, Z. et al. Genome Biol. 11, R31 (2010).
Montange, R.K. et al. J. Mol. Biol. 396, 761–772 (2010).
Dieckmann, T., Suzuki, E., Nakamura, G.K. & Feigon, J. RNA 2, 628–640 (1996).
Block, K.F., Hammond, M.C. & Breaker, R.R. J. Bacteriol. 192, 3983–3989 (2010).
McTaggart, J.S., Clark, R.H. & Ashcroft, F.M. J. Physiol. (Lond.) 588, 3201–3209 (2010).
Dennis, P.B. et al. Science 294, 1102–1105 (2001).
McCabe, B.C. & Gollnick, P. J. Bacteriol. 186, 5157–5159 (2004).
Kato-Yamada, Y. FEBS Lett. 579, 6875–6878 (2005).
Gaal, T., Bartlett, M.S., Ross, W., Turnbough, C.L. Jr. & Gourse, R.L. Science 278, 2092–2097 (1997).
Regulski, E.E. & Breaker, R.R. Methods Mol. Biol. 419, 53–67 (2008).
Montange, R.K. & Batey, R.T. Annu. Rev. Biophys. 37, 117–133 (2008).
Watson, P.Y. & Fedor, M.J. Nat. Struct. Mol. Biol. 18, 359–363 (2011).
Noeske, J. et al. Proc. Natl. Acad. Sci. USA 102, 1372–1377 (2005).
Allenby, N.E. et al. J. Bacteriol. 187, 8063–8080 (2005).
Guffanti, A.A., Clejan, S., Falk, L.H., Hicks, D.B. & Krulwich, T.A. J. Bacteriol. 169, 4469–4478 (1987).
Barrick, J.E. et al. Proc. Natl. Acad. Sci. USA 101, 6421–6426 (2004).
Holtmann, G., Bakker, E.P., Uozumi, N. & Bremer, E. J. Bacteriol. 185, 1289–1298 (2003).
Lee, E.J. & Groisman, E.A. Nature 486, 271–275 (2012).
Acknowledgements
We acknowledge J. Viladoms for thoughtful conversations and critical reading of this manuscript. We thank the groups of E. Winzeler and P. Sun for access to qPCR instruments and the Skaggs Institute for Chemical Biology for graduate fellowship support of P.Y. Watson.
Author information
Authors and Affiliations
Contributions
P.Y.W. and M.J.F. conceived of and designed experiments, analyzed the data and prepared the manuscript. P.Y.W. conducted the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 1423 kb)
Rights and permissions
About this article
Cite this article
Watson, P., Fedor, M. The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis. Nat Chem Biol 8, 963–965 (2012). https://doi.org/10.1038/nchembio.1095
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1095
This article is cited by
-
ATP dynamic regeneration strategy for enhancing co-production of glutathione and S-adenosylmethionine in Escherichia coli
Biotechnology Letters (2020)
-
Recent advances and future trends of riboswitches: attractive regulatory tools
World Journal of Microbiology and Biotechnology (2018)
-
Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage
Nature Chemical Biology (2016)
-
Secondary structural entropy in RNA switch (Riboswitch) identification
BMC Bioinformatics (2015)
-
Greatest hits
Nature Chemical Biology (2015)