Abstract
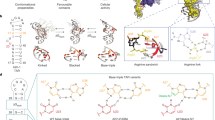
Current approaches used to identify protein-binding small molecules are not suited for identifying small molecules that can bind emerging RNA drug targets. By docking small molecules onto an RNA dynamic ensemble constructed by combining NMR spectroscopy and computational molecular dynamics, we virtually screened small molecules that target the entire structure landscape of the transactivation response element (TAR) from HIV type 1 (HIV-1). We quantitatively predict binding energies for small molecules that bind different RNA conformations and report the de novo discovery of six compounds that bind TAR with high affinity and inhibit its interaction with a Tat peptide in vitro (Ki values of 710 nM–169 μM). One compound binds HIV-1 TAR with marked selectivity and inhibits Tat-mediated activation of the HIV-1 long terminal repeat by 81% in T-cell lines and HIV replication in an HIV-1 indicator cell line (IC50 ∼23.1 μM).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cooper, T.A., Wan, L. & Dreyfuss, G. RNA and disease. Cell 136, 777–793 (2009).
Parsons, J. et al. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat. Chem. Biol. 5, 823–825 (2009).
Blount, K.F. & Breaker, R.R. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 24, 1558–1564 (2006).
Thomas, J.R. & Hergenrother, P.J. Targeting RNA with small molecules. Chem. Rev. 108, 1171–1224 (2008).
Kuntz, I.D. Structure-based strategies for drug design and discovery. Science 257, 1078–1082 (1992).
Filikov, A.V. et al. Identification of ligands for RNA targets via structure-based virtual screening: HIV-1 TAR. J. Comput. Aided Mol. Des. 14, 593–610 (2000).
Hermann, T. Rational ligand design for RNA: the role of static structure and conformational flexibility in target recognition. Biochimie 84, 869–875 (2002).
Cruz, J.A. & Westhof, E. The dynamic landscapes of RNA architecture. Cell 136, 604–609 (2009).
Fulle, S. & Gohlke, H. Molecular recognition of RNA: challenges for modelling interactions and plasticity. J. Mol. Recognit. 23, 220–231 (2010).
Zhang, Q., Sun, X., Watt, E.D. & Al-Hashimi, H.M. Resolving the motional modes that code for RNA adaptation. Science 311, 653–656 (2006).
Frank, A.T., Stelzer, A.C., Al-Hashimi, H.M. & Andricioaei, I. Constructing RNA dynamical ensembles by combining MD and motionally decoupled NMR RDCs: new insights into RNA dynamics and adaptive ligand recognition. Nucleic Acids Res. 37, 3670–3679 (2009).
Puglisi, J.D., Tan, R., Calnan, B.J., Frankel, A.D. & Williamson, J.R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 257, 76–80 (1992).
Williamson, J.R. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 7, 834–837 (2000).
Leulliot, N. & Varani, G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry 40, 7947–7956 (2001).
Zhang, Q., Stelzer, A.C., Fisher, C.K. & Al-Hashimi, H.M. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 450, 1263–1267 (2007).
Latham, M.P., Zimmermann, G.R. & Pardi, A. NMR chemical exchange as a probe for ligand-binding kinetics in a theophylline-binding RNA aptamer. J. Am. Chem. Soc. 131, 5052–5053 (2009).
Vaiana, A.C. & Sanbonmatsu, K.Y. Stochastic gating and drug-ribosome interactions. J. Mol. Biol. 386, 648–661 (2009).
Chen, Y., Campbell, S.L. & Dokholyan, N.V. Deciphering protein dynamics from NMR data using explicit structure sampling and selection. Biophys. J. 93, 2300–2306 (2007).
Clore, G.M. & Schwieters, C.D. Amplitudes of protein backbone dynamics and correlated motions in a small alpha/beta protein: correspondence of dipolar coupling and heteronuclear relaxation measurements. Biochemistry 43, 10678–10691 (2004).
Abagyan, R., Totrov, M. & Kuznetsov, D. ICM—a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 15, 488–506 (1994).
Lang, P.T. et al. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA 15, 1219–1230 (2009).
Cheng, T., Li, X., Li, Y., Liu, Z. & Wang, R. Comparative assessment of scoring functions on a diverse test set. J. Chem. Inf. Model. 49, 1079–1093 (2009).
Guilbert, C. & James, T.L. Docking to RNA via root-mean-square-deviation-driven energy minimization with flexible ligands and flexible targets. J. Chem. Inf. Model. 48, 1257–1268 (2008).
Ippolito, J.A. & Steitz, T.A. A 1.3-Å resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc. Natl. Acad. Sci. USA 95, 9819–9824 (1998).
Aboul-ela, F., Karn, J. & Varani, G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 24, 3974–3981 (1996).
Yang, M. Discoveries of Tat-TAR interaction inhibitors for HIV-1. Curr. Drug Targets Infect. Disord. 5, 433–444 (2005).
Bradrick, T.D. & Marino, J.P. Ligand-induced changes in 2-aminopurine fluorescence as a probe for small molecule binding to HIV-1 TAR RNA. RNA 10, 1459–1468 (2004).
Matsumoto, C., Hamasaki, K., Mihara, H. & Ueno, A. A high-throughput screening utilizing intramolecular fluorescence resonance energy transfer for the discovery of the molecules that bind HIV-1 TAR RNA specifically. Bioorg. Med. Chem. Lett. 10, 1857–1861 (2000).
Davidson, A., Patora-Komisarska, K., Robinson, J.A. & Varani, G. Essential structural requirements for specific recognition of HIV TAR RNA by peptide mimetics of Tat protein. Nucleic Acids Res. 39, 248–256 (2011).
Davidson, A. et al. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc. Natl. Acad. Sci. USA 106, 11931–11936 (2009).
White, R.J. & Durr, F.E. Development of mitoxantrone. Invest. New Drugs 3, 85–93 (1985).
Parolin, C. et al. New anti-human immunodeficiency virus type 1 6-aminoquinolones: mechanism of action. Antimicrob. Agents Chemother. 47, 889–896 (2003).
Blount, K.F., Tor, Y., Hamasaki, K. & Ueno, A. Using pyrene-labeled HIV-1 TAR to measure RNA-small molecule binding aminoglycoside antibiotics, neamine and its derivatives as potent inhibitors for the RNA-protein interactions derived from HIV-1 activators. Nucleic Acids Res. 31, 5490–5500 (2003).
DeJong, E.S., Chang, C.E., Gilson, M.K. & Marino, J.P. Proflavine acts as a Rev inhibitor by targeting the high-affinity Rev binding site of the Rev responsive element of HIV-1. Biochemistry 42, 8035–8046 (2003).
Kaul, M., Barbieri, C.M. & Pilch, D.S. Fluorescence-based approach for detecting and characterizing anti biotic-induced conformational changes in ribosomal RNA: Comparing aminoglycoside binding to prokaryotic and eukaryotic ribosomal RNA sequences. J. Am. Chem. Soc. 126, 3447–3453 (2004).
Stelzer, A.C., Kratz, J.D., Zhang, Q. & Al-Hashimi, H.M. RNA dynamics by design: biasing ensembles towards the ligand-bound state. Angew. Chem. Int. Ed. Engl. 49, 5731–5733 (2010).
Lapidot, A., Berchanski, A. & Borkow, G. Insight into the mechanisms of aminoglycoside derivatives interaction with HIV-1 entry steps and viral gene transcription. FEBS J. 275, 5236–5257 (2008).
Lapidot, A., Vijayabaskar, V., Litovchick, A., Yu, J.G. & James, T.L. Structure-activity relationships of amino glyco side-arginine conjugates that bind HIV-1 RNAs as determined by fluorescence and NMR spectroscopy. FEBS Lett. 577, 415–421 (2004).
Faber, C., Sticht, H., Schweimer, K. & Rosch, P. Structural rearrangements of HIV-1 Tat-responsive RNA upon binding of neomycin B. J. Biol. Chem. 275, 20660–20666 (2000).
Cabrera, C. et al. Anti-HIV activity of a novel aminoglycoside-arginine conjugate. Antiviral Res. 53, 1–8 (2002).
Blount, K.F., Zhao, F., Hermann, T. & Tor, Y. Conformational constraint as a means for understanding RNA-aminoglycoside specificity. J. Am. Chem. Soc. 127, 9818–9829 (2005).
Boehr, D.D., Nussinov, R. & Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR 6, 277–293 (1995).
Acknowledgements
We thank A.V. Kurochkin for NMR expertise, and we thank the Michigan Economic Development Cooperation and the Michigan Technology Tri-Corridor for support of the purchase of a 600-MHz spectrometer. This work was supported by the US National Institutes of Health (R01 AI066975-01 and R01 CA144043), the US National Science Foundation (NSF Career Award CHE-0918817) and an NSF Graduate Research Fellowship for A.C.S. and A.T.F.
Author information
Authors and Affiliations
Contributions
H.M.A.-H. and A.C.S. conceived the docking approach; A.T.F., I.A., A.C.S. and H.M.A.-H. developed the approach for constructing RNA dynamic structure ensembles; A.C.S., with input from A.T.F. and J.D.K., performed the docking simulations; A.C.S., J.D.K. and J.L. preformed the in vitro fluorescence assays and NMR experiments; M.D.S., M.J.G.-H and D.M.M. carried out the transfection and viral-replication assays; H.M.A.-H., A.C.S., A.T.F., J.D.K., I.A. and D.M.M. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
H.M.A.-H. is an advisor to and holds an ownership interest in Nymirum, an RNA-based drug-discovery company. A.C.S. completed this work as part of his dissertation and is now employed by Nymirum. The research reported in this article was performed by the University of Michigan faculty and students and was funded by a US National Institutes of Health contract to H.M.A.-H.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Methods (PDF 3462 kb)
Supplementary Data
Supplementary Data (XLSX 13 kb)
Rights and permissions
About this article
Cite this article
Stelzer, A., Frank, A., Kratz, J. et al. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol 7, 553–559 (2011). https://doi.org/10.1038/nchembio.596
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.596
This article is cited by
-
Structural and dynamic mechanisms for coupled folding and tRNA recognition of a translational T-box riboswitch
Nature Communications (2023)
-
Targeting RNA structures with small molecules
Nature Reviews Drug Discovery (2022)
-
Targeting Xist with compounds that disrupt RNA structure and X inactivation
Nature (2022)
-
Design of a small molecule that stimulates vascular endothelial growth factor A enabled by screening RNA fold–small molecule interactions
Nature Chemistry (2020)
-
IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex
Nature Communications (2020)