Abstract
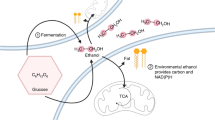
The interaction between fermentation-respiration switch (FrsA) protein and glucose-specific enzyme IIAGlc increases glucose fermentation under oxygen-limited conditions. We show that FrsA converts pyruvate to acetaldehyde and carbon dioxide in a cofactor-independent manner and that its pyruvate decarboxylation activity is enhanced by the dephosphorylated form of IIAGlc (d-IIAGlc). Crystal structures of FrsA and its complex with d-IIAGlc revealed residues required for catalysis as well as the structural basis for the activation by d-IIAGlc.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Saier, M.H. Jr., Novotny, M.J., Comeau-Fuhrman, D., Osumi, T. & Desai, J.D. J. Bacteriol. 155, 1351–1357 (1983).
Peterkofsky, A. et al. Prog. Nucleic Acid Res. Mol. Biol. 44, 31–65 (1993).
Nam, T.W. et al. Proc. Natl. Acad. Sci. USA 105, 3751–3756 (2008).
Seok, Y.J. et al. J. Biol. Chem. 272, 26511–26521 (1997).
Kim, Y.J. et al. FEBS Lett. 584, 4537–4544 (2010).
Koo, B.M. et al. J. Biol. Chem. 279, 31613–31621 (2004).
Ollis, D.L. et al. Protein Eng. 5, 197–211 (1992).
Lee, S.J. et al. J. Biol. Chem. 278, 44552–44559 (2003).
Wu, N., Mo, Y., Gao, J. & Pai, E.F. Proc. Natl. Acad. Sci. USA 97, 2017–2022 (2000).
Appleby, T.C., Kinsland, C., Begley, T.P. & Ealick, S.E. Proc. Natl. Acad. Sci. USA 97, 2005–2010 (2000).
Harris, P., Navarro Poulsen, J.C., Jensen, K.F. & Larsen, S. Biochemistry 39, 4217–4224 (2000).
Miller, B.G., Hassell, A.M., Wolfenden, R., Milburn, M.V. & Short, S.A. Proc. Natl. Acad. Sci. USA 97, 2011–2016 (2000).
Chan, K.K. et al. Biochemistry 48, 5518–5531 (2009).
Cendron, L. et al. J. Biol. Chem. 282, 18182–18189 (2007).
Kim, K., Park, J. & Rhee, S. J. Biol. Chem. 282, 23457–23464 (2007).
Jencks, W.P. Adv. Enzymol. 43, 219–410 (1975).
Erecińska, M. & Silver, I.A. Respir. Physiol. 128, 263–276 (2001).
Chun, C.D., Liu, O.W. & Madhani, H.D. PLoS Pathog. 3, e22 (2007).
Arjunan, P. et al. J. Mol. Biol. 256, 590–600 (1996).
Dobritzsch, D., Konig, S., Schneider, G. & Lu, G. J. Biol. Chem. 273, 20196–20204 (1998).
Dyda, F. et al. Biochemistry 32, 6165–6170 (1993).
Stephenson, M.P. & Dawes, E.A. J. Gen. Microbiol. 69, 331–343 (1971).
Hogema, B.M. et al. Mol. Microbiol. 30, 487–498 (1998).
Bringer-Meyer, S., Schimz, K.L. & Sahm, H. Arch. Microbiol. 146, 105–110 (1986).
Sergienko, E.A. & Jordan, F. Biochemistry 40, 7369–7381 (2001).
Acknowledgements
We are grateful to Y.-J. Lee, H.-S. Lee, S.J. Chung and J.K. Lee for comments on the manuscript. This work was supported by the Marine and Extreme Genome Research Center program of Ministry of Land, Transport and Maritime Affairs, the Mid-career Researcher Program through National Research Foundation grant funded by the Ministry of Education, Science and Technology (2009-0092822), a KORDI in-house program (PE98513) and the Development of Biohydrogen Production Technology Using Hyperthermophilic Archaea program of Ministry of Land, Transport and Maritime Affairs.
Author information
Authors and Affiliations
Contributions
S.-S.C. and K.-H.L. designed the project, analyzed data and wrote the manuscript. K.-J.L., C.-S.J., Y.J.A., H.-J.L., S.-J.P. and S.-S.C. performed experiments. Y.-J.S. gave advice on the design of this project and the NMR experiments. P.K. gave advice on the biochemical experiments. J.-H.L. contributed analytic tools.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 4237 kb)
Rights and permissions
About this article
Cite this article
Lee, KJ., Jeong, CS., An, Y. et al. FrsA functions as a cofactor-independent decarboxylase to control metabolic flux. Nat Chem Biol 7, 434–436 (2011). https://doi.org/10.1038/nchembio.589
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.589
This article is cited by
-
Vibrio vulnificus induces the death of a major bacterial species in the mouse gut via cyclo-Phe-Pro
Microbiome (2021)
-
Role of DegQ in differential stability of flagellin subunits in Vibrio vulnificus
npj Biofilms and Microbiomes (2021)
-
Role of AcsR in expression of the acetyl-CoA synthetase gene in Vibrio vulnificus
BMC Microbiology (2015)
-
Structure and functional characterization of pyruvate decarboxylase from Gluconacetobacter diazotrophicus
BMC Structural Biology (2014)
-
VvpM, an extracellular metalloprotease of Vibrio vulnificus, induces apoptotic death of human cells
Journal of Microbiology (2014)