Abstract
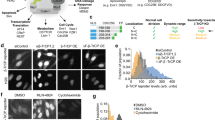
Fluorescent biosensors for living cells currently require laborious optimization and a unique design for each target. They are limited by the availability of naturally occurring ligands with appropriate target specificity. Here we describe a biosensor based on an engineered fibronectin monobody scaffold that can be tailored to bind different targets via high-throughput screening. We made this Src-family kinase (SFK) biosensor by derivatizing a monobody specific for activated SFKs with a bright dye whose fluorescence increases upon target binding. We identified sites for dye attachment and changes to eliminate vesiculation in living cells, providing a generalizable scaffold for biosensor production. This approach minimizes cell perturbation because it senses endogenous, unmodified target, and because sensitivity is enhanced by direct dye excitation. Automated correlation of cell velocities and SFK activity revealed that SFKs are activated specifically during protrusion. Activity correlates with velocity, and peaks 1–2 μm from the leading edge.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Change history
15 June 2012
In the version of this article initially published, the residue numbers indicated in the y-axis labels in Figure 3c were incorrectly written as 24, 53, 55 and 53 from the origin but should have read 24, 52, 53 and 55. The error has been corrected for the PDF and HTML versions of this article.
References
Nalbant, P., Hodgson, L., Kraynov, V., Toutchkine, A. & Hahn, K.M. Activation of endogenous Cdc42 visualized in living cells. Science 305, 1615–1619 (2004).
Toutchkine, A., Kraynov, V. & Hahn, K. Solvent-sensitive dyes to report protein conformational changes in living cells. J. Am. Chem. Soc. 125, 4132–4145 (2003).
Toutchkine, A., Nguyen, D.V. & Hahn, K.M. Merocyanine dyes with improved photostability. Org. Lett. 9, 2775–2777 (2007).
Toutchkine, A., Nguyen, D.V. & Hahn, K.M. Simple one-pot preparation of water-soluble, cysteine-reactive cyanine and merocyanine dyes for biological imaging. Bioconjug. Chem. 18, 1344–1348 (2007).
Koide, A., Bailey, C.W., Huang, X. & Koide, S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 284, 1141–1151 (1998).
Karatan, E. et al. Molecular recognition properties of FN3 monobodies that bind the Src SH3 domain. Chem. Biol. 11, 835–844 (2004).
Koide, A., Abbatiello, S., Rothgery, L. & Koide, S. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc. Natl. Acad. Sci. USA 99, 1253–1258 (2002).
Sidhu, S.S. & Koide, S. Phage display for engineering and analyzing protein interaction interfaces. Curr. Opin. Struct. Biol. 17, 481–487 (2007).
Parsons, S.J. & Parsons, J.T. Src family kinases, key regulators of signal transduction. Oncogene 23, 7906–7909 (2004).
Thomas, S.M. & Brugge, J.S. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 13, 513–609 (1997).
Playford, M.P. & Schaller, M.D. The interplay between Src and integrins in normal and tumor biology. Oncogene 23, 7928–7946 (2004).
Bromann, P.A., Korkaya, H. & Courtneidge, S.A. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene 23, 7957–7968 (2004).
Ouyang, M., Sun, J., Chien, S. & Wang, Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc. Natl. Acad. Sci. USA 105, 14353–14358 (2008).
Wang, Y. et al. Visualizing the mechanical activation of Src. Nature 434, 1040–1045 (2005).
Ting, A.Y., Kain, K.H., Klemke, R.L. & Tsien, R.Y. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc. Natl. Acad. Sci. USA 98, 15003–15008 (2001).
Cowan-Jacob, S.W. et al. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure 13, 861–871 (2005).
Gardner, O.S., Dewar, B.J., Earp, H.S., Samet, J.M. & Graves, L.M. Dependence of peroxisome proliferator-activated receptor ligand-induced mitogen-activated protein kinase signaling on epidermal growth factor receptor transactivation. J. Biol. Chem. 278, 46261–46269 (2003).
Dewar, B.J. et al. Capacitative calcium entry contributes to the differential transactivation of the epidermal growth factor receptor in response to thiazolidinediones. Mol. Pharmacol. 72, 1146–1156 (2007).
Rizzo, M.A., Springer, G.H., Granada, B. & Piston, D.W. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22, 445–449 (2004).
Fosbrink, M., Aye-Han, N.N., Cheong, R., Levchenko, A. & Zhang, J. Visualization of JNK activity dynamics with a genetically encoded fluorescent biosensor. Proc. Natl. Acad. Sci. USA 107, 5459–5464 (2010).
Pertz, O., Hodgson, L., Klemke, R.L. & Hahn, K.M. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440, 1069–1072 (2006).
Glick, D., Barth, S. & Macleod, K.F. Autophagy: cellular and molecular mechanisms. J. Pathol. 221, 3–12 (2010).
Reis, R.C., Sorgine, M.H. & Coelho-Sampaio, T. A novel methodology for the investigation of intracellular proteolytic processing in intact cells. Eur. J. Cell Biol. 75, 192–197 (1998).
Panchuk-Voloshina, N. et al. Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J. Histochem. Cytochem. 47, 1179–1188 (1999).
Hodgson, L., Shen, F. & Hahn, K. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr. Protoc. Cell Biol. 46, 14.11.1–14.11.26 (2010).
Bright, G.R., Fisher, G.W., Rogowska, J. & Taylor, D.L. Fluorescence ratio imaging microscopy. Methods Cell Biol. 30, 157–192 (1989).
Buccione, R., Orth, J.D. & McNiven, M.A. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 5, 647–657 (2004).
Veracini, L. et al. Two functionally distinct pools of Src kinases for PDGF receptor signalling. Biochem. Soc. Trans. 33, 1313–1315 (2005).
Veracini, L. et al. Two distinct pools of Src family tyrosine kinases regulate PDGF-induced DNA synthesis and actin dorsal ruffles. J. Cell Sci. 119, 2921–2934 (2006).
Sandilands, E., Brunton, V.G. & Frame, M.C. The membrane targeting and spatial activation of Src, Yes and Fyn is influenced by palmitoylation and distinct RhoB/RhoD endosome requirements. J. Cell Sci. 120, 2555–2564 (2007).
Sandilands, E. et al. RhoB and actin polymerization coordinate Src activation with endosome-mediated delivery to the membrane. Dev. Cell 7, 855–869 (2004).
Cary, L.A., Klinghoffer, R.A., Sachsenmaier, C. & Cooper, J.A. SRC catalytic but not scaffolding function is needed for integrin-regulated tyrosine phosphorylation, cell migration, and cell spreading. Mol. Cell. Biol. 22, 2427–2440 (2002).
Fincham, V.J. & Frame, M.C. The catalytic activity of Src is dispensable for translocation to focal adhesions but controls the turnover of these structures during cell motility. EMBO J. 17, 81–92 (1998).
Frame, M.C., Fincham, V.J., Carragher, N.O. & Wyke, J.A. v-Src's hold over actin and cell adhesions. Nat. Rev. Mol. Cell Biol. 3, 233–245 (2002).
Machacek, M. & Danuser, G. Morphodynamic profiling of protrusion phenotypes. Biophys. J. 90, 1439–1452 (2006).
Machacek, M. et al. Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103 (2009).
Renard, M. et al. Knowledge-based design of reagentless fluorescent biosensors from recombinant antibodies. J. Mol. Biol. 318, 429–442 (2002).
Koide, A., Jordan, M.R., Horner, S.R., Batori, V. & Koide, S. Stabilization of a fibronectin type III domain by the removal of unfavorable electrostatic interactions on the protein surface. Biochemistry 40, 10326–10333 (2001).
Kraynov, V.S. et al. Localized Rac activation dynamics visualized in living cells. Science 290, 333–337 (2000).
Martinez-Quiles, N., Ho, H.Y., Kirschner, M.W., Ramesh, N. & Geha, R.S. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol. Cell. Biol. 24, 5269–5280 (2004).
Cory, G.O., Garg, R., Cramer, R. & Ridley, A.J. Phosphorylation of tyrosine 291 enhances the ability of WASp to stimulate actin polymerization and filopodium formation. Wiskott-Aldrich Syndrome protein. J. Biol. Chem. 277, 45115–45121 (2002).
Suetsugu, S. et al. Sustained activation of N-WASP through phosphorylation is essential for neurite extension. Dev. Cell 3, 645–658 (2002).
Plattner, R., Kadlec, L., DeMali, K.A., Kazlauskas, A. & Pendergast, A.M. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 13, 2400–2411 (1999).
Sini, P., Cannas, A., Koleske, A.J., Di Fiore, P.P. & Scita, G. Abl-dependent tyrosine phosphorylation of Sos-1 mediates growth-factor-induced Rac activation. Nat. Cell Biol. 6, 268–274 (2004).
Yang, L., Kowalski, J.R., Zhan, X., Thomas, S.M. & Luscinskas, F.W. Endothelial cell cortactin phosphorylation by Src contributes to polymorphonuclear leukocyte transmigration in vitro. Circ. Res. 98, 394–402 (2006).
Bouton, A.H., Riggins, R.B. & Bruce-Staskal, P.J. Functions of the adapter protein Cas: signal convergence and the determination of cellular responses. Oncogene 20, 6448–6458 (2001).
Polte, T.R. & Hanks, S.K. Complexes of focal adhesion kinase (FAK) and Crk-associated substrate (p130(Cas)) are elevated in cytoskeleton-associated fractions following adhesion and Src transformation. Requirements for Src kinase activity and FAK proline-rich motifs. J. Biol. Chem. 272, 5501–5509 (1997).
DerMardirossian, C., Rocklin, G., Seo, J.Y. & Bokoch, G.M. Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol. Biol. Cell 17, 4760–4768 (2006).
Timpson, P., Jones, G.E., Frame, M.C. & Brunton, V.G. Coordination of cell polarization and migration by the Rho family GTPases requires Src tyrosine kinase activity. Curr. Biol. 11, 1836–1846 (2001).
Rossman, K.L., Der, C.J. & Sondek, J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6, 167–180 (2005).
Acknowledgements
We thank C. MacNevin for assistance with dye synthesis, F. Shen for help with imaging studies, D. Renfrew for assistance with computational modeling, A. Nguyen for technical assistance and B. Clarke for administrative assistance. We gratefully acknowledge funding from the American Heart Association (A.G.) and the US National Institutes of Health (GM GM082288 and GM057464 to K.M.H.).
Author information
Authors and Affiliations
Contributions
A.G. developed the biosensors, with help from B.D. and L.M.G. on phosphorylation assays. E.V. carried out live-cell imaging studies, assisted by A.G. R.A. and T.E. developed the image analysis algorithms and carried out automated analysis of SFK activity, with help from J.W. S.L. and B.K. modeled dye-protein interactions. D.G. synthesized the dyes. B.K.K. provided protein constructs and input regarding FN3 screening and structure. K.M.H. conceived the study, directed the research and wrote the final manuscript based on contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results and Supplementary Methods (PDF 1299 kb)
Supplementary Video 1
Src activation in fibroblasts with PDGF stimulation (AVI 5283 kb)
Supplementary Video 2
Src activation at the cell edge in PDGF stimulated fibroblasts (AVI 482 kb)
Supplementary Video 3
Src activation in randomly migrating epithelial cells (AVI 5024 kb)
Supplementary Video 4
Src activation at the cell edge and the effect of inhibitor (AVI 865 kb)
Supplementary Video 5
Merobody sensor is sensitive to Src activity. (AVI 4690 kb)
Supplementary Video 6
Src merobody sensor localization at the cell edge is dependent on Src activity (AVI 649 kb)
Rights and permissions
About this article
Cite this article
Gulyani, A., Vitriol, E., Allen, R. et al. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nat Chem Biol 7, 437–444 (2011). https://doi.org/10.1038/nchembio.585
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.585
This article is cited by
-
Subcellular and Dynamic Coordination between Src Activity and Cell Protrusion in Microenvironment
Scientific Reports (2015)
-
Characterization of Monobody Scaffold Interactions with Ligand via Force Spectroscopy and Steered Molecular Dynamics
Scientific Reports (2015)
-
Erratum: Corrigendum: A biosensor generated via high-throughput screening quantifies cell edge Src dynamics
Nature Chemical Biology (2012)
-
Imaging the coordination of multiple signalling activities in living cells
Nature Reviews Molecular Cell Biology (2011)