Abstract
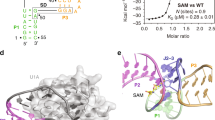
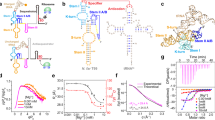
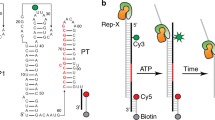
Riboswitches are gene regulation elements in mRNA that function by specifically responding to metabolites. Although the metabolite-bound states of riboswitches have proven amenable to structure determination efforts, knowledge of the structural features of riboswitches in their ligand-free forms and their ligand-response mechanisms giving rise to regulatory control is lacking. Here we explore the ligand-induced folding process of the S-adenosylmethionine type II (SAM-II) riboswitch using chemical and biophysical methods, including NMR and fluorescence spectroscopy, and single-molecule fluorescence imaging. The data reveal that the unliganded SAM-II riboswitch is dynamic in nature, in that its stem-loop element becomes engaged in a pseudoknot fold through base-pairing with nucleosides in the 3′ overhang containing the Shine-Dalgarno sequence. Although the pseudoknot structure is highly transient in the absence of its ligand, S-adenosylmethionine (SAM), it becomes conformationally restrained upon ligand recognition, through a conformational capture mechanism. These insights provide a molecular understanding of riboswitch dynamics that shed new light on the mechanism of riboswitch-mediated translational regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Roth, A. & Breaker, R.R. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 78, 305–334 (2009).
Montange, R.K. & Batey, R.T. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys 37, 117–133 (2008).
Blouin, S., Mulhbacher, J., Penedo, J.C. & Lafontaine, D.A. Riboswitches: ancient and promising genetic regulators. ChemBioChem 10, 400–416 (2009).
Serganov, A. & Patel, D.J. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 8, 776–790 (2007).
Schwalbe, H., Buck, J., Fürtig, B., Noeske, J. & Wöhnert, J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew. Chem. Int. Edn. Engl. 46, 1212–1219 (2007).
Nudler, E. & Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 29, 11–17 (2004).
Serganov, A. Determination of riboswitch structures: light at the end of the tunnel? RNA Biol. 7, 98–103 (2010).
Gilbert, S.D., Rambo, R.P., Van Tyne, D. & Batey, R.T. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 15, 177–182 (2008).
Garst, A.D. & Batey, R.T. A switch in time: detailing the life of a riboswitch. Biochim. Biophys. Acta 1789, 584–591 (2009).
Stoddard, C.D. et al. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18, 787–797 (2010).
Baird, N.J. & Ferré-D'Amaré, A.R. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16, 598–609 (2010).
Edwards, A.L., Reyes, F.E., Héroux, A. & Batey, R.T. Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA 16, 2144–2155 (2010).
Bosshard, H.R. Molecular recognition by induced fit: how fit is the concept? News Physiol. Sci. 16, 171–173 (2001).
Weikl, T.R. & von Deuster, C. Selected-fit versus induced-fit protein binding: Kinetic differences and mutational analysis. Proteins 75, 104–110 (2009).
Noeske, J. et al. Interplay of 'induced fit' and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 35, 572–583 (2007).
Boehr, D.D., Nussinov, R. & Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009).
Leulliot, N. & Varani, G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry 40, 7947–7956 (2001).
Duchardt-Ferner, E. et al. Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch. Angew. Chem. Int. Edn Engl. 49, 6216–6219 (2010).
Stelzer, A.C., Kratz, J.D., Zhang, Q. & Al-Hashimi, H.M. RNA dynamics by design: biasing ensembles towards the ligand-bound state. Angew. Chem. Int. Edn Engl. 49, 5731–5733 (2010).
Wang, J.X. & Breaker, R.R. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem. Cell Biol. 86, 157–168 (2008).
Poiata, E., Meyer, M.M., Ames, T.D. & Breaker, R.R. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA 15, 2046–2056 (2009).
Corbino, K.A. et al. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 6, R70 (2005).
Rieder, U., Lang, K., Kreutz, C., Polacek, N. & Micura, R. Evidence for pseudoknot formation of class I preQ1 riboswitch aptamers. ChemBioChem 10, 1141–1144 (2009).
Puffer, B. et al. 5-Fluoro pyrimidines: labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy. Nucleic Acids Res. 37, 7728–7740 (2009).
Rieder, U., Kreutz, C. & Micura, R. Folding of a transcriptionally acting preQ1 riboswitch. Proc. Natl. Acad. Sci. USA 107, 10804–10809 (2010).
Lang, K. & Micura, R. The preparation of site-specifically modified riboswitch domains as an example for enzymatic ligation of chemically synthesized RNA fragments. Nat. Protoc. 3, 1457–1466 (2008).
Alemán, E.A., Lamichhane, R. & Rueda, D. Exploring RNA folding one molecule at a time. Curr. Opin. Chem. Biol. 12, 647–654 (2008).
Lemay, J.F., Penedo, J.C., Tremblay, R., Lilley, D.M. & Lafontaine, D.A. Folding of the adenine riboswitch. Chem. Biol. 13, 857–868 (2006).
Brenner, M.D., Scanlan, M.S., Nahas, M.K., Ha, T. & Silverman, S.K. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry 49, 1596–1605 (2010).
Munro, J.B., Altman, R.B., O'Connor, N. & Blanchard, S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell 25, 505–517 (2007).
Lang, K., Rieder, R. & Micura, R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 35, 5370–5378 (2007).
Rieder, R., Lang, K., Graber, D. & Micura, R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem 8, 896–902 (2007).
Kelley, J.M. & Hamelberg, D. Atomistic basis for the on-off signaling mechanism in SAM-II riboswitch. Nucleic Acids Res. 38, 1392–1400 (2010).
Mortimer, S.A. & Weeks, K.M. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 129, 4144–4145 (2007).
Weeks, K.M. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 20, 295–304 (2010).
Staple, D.W. & Butcher, S.E. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 3, e213 (2005).
Serganov, A., Ennifar, E., Portier, C., Ehresmann, B. & Ehresmann, C. Do mRNA and rRNA binding sites of E.coli ribosomal protein S15 share common structural determinants? J. Mol. Biol. 320, 963–978 (2002).
Munro, J.B., Wasserman, M.R., Altman, R.B., Wang, L. & Blanchard, S.C. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat. Struct. Mol. Biol. 17, 1470–1477 (2010).
Al-Hashimi, H.M. & Walter, N.G. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 18, 321–329 (2008).
Zhang, Q., Stelzer, A.C., Fisher, C.K. & Al-Hashimi, H.M. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 450, 1263–1267 (2007).
Frank, A.T., Stelzer, A.C., Al-Hashimi, H.M. & Andricioaei, I. Constructing RNA dynamical ensembles by combining MD and motionally decoupled NMR RDCs: new insights into RNA dynamics and adaptive ligand recognition. Nucleic Acids Res. 37, 3670–3679 (2009).
Lu, C. et al. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J. Mol. Biol. 404, 803–818 (2010).
Wilson, R.C. et al. Tuning riboswitch regulation through conformational selection. J. Mol. Biol. 4, 926–938 (2011).
Ottink, O.M. et al. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA 13, 2202–2212 (2007).
Noeske, J., Schwalbe, H. & Wöhnert, J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 35, 5262–5273 (2007).
Lee, M.K., Gal, M., Frydman, L. & Varani, G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc. Natl. Acad. Sci. USA 107, 9192–9197 (2010).
Hermann, T. & Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 287, 820–825 (2000).
Dave, R., Terry, D.S., Munro, J.B. & Blanchard, S.C. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96, 2371–2381 (2009).
Qin, F. & Li, L. Model-based fitting of single-channel dwell-time distributions. Biophys. J. 87, 1657–1671 (2004).
Lescoute, A. & Westhof, E. The interaction networks of structured RNAs. Nucleic Acids Res. 34, 6587–6604 (2006).
Acknowledgements
We thank D. Lafontaine for discussions on labeling of riboswitches for smFRET experiments; K. Lang (University of Innsbruck; presently at Medical Research Council, Cambridge) for a previous synthesis of 15N-labeled cytidine phosphoramidite; C. Kreutz and M. Tollinger for NMR assistance; D.S. Terry for contributions to the development and implementation of the single-molecule imaging platform and data analysis software used; and M. Soulière for critical reading of the manuscript. This work was funded by the Austrian Science Foundation FWF (I317 and P21641 to R.M.) and the Austrian Ministry of Science and Research (GenAU project consortium 'non-coding RNAs' P0726-012-012 to R.M.). S.C.B. was supported by the Irma T. Hirschl/Monique Weill-Caulier Trust.
Author information
Authors and Affiliations
Contributions
A.H. performed RNA synthesis, labeling, enzymatic ligations and ensemble fluorescence spectroscopic measurements. S.C.B. performed smFRET measurements. U.R. performed NMR experiments. M.A. synthesized 5-fluorocytidine phosphoramidite for RNA synthesis. R.M., S.C.B. and A.H. analyzed the data. R.M. designed the study and wrote the paper (together with S.C.B. and A.H.).
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Figures 1–7 and Supplementary Table 1 (PDF 1781 kb)
Rights and permissions
About this article
Cite this article
Haller, A., Rieder, U., Aigner, M. et al. Conformational capture of the SAM-II riboswitch. Nat Chem Biol 7, 393–400 (2011). https://doi.org/10.1038/nchembio.562
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.562
This article is cited by
-
Observation of structural switch in nascent SAM-VI riboswitch during transcription at single-nucleotide and single-molecule resolution
Nature Communications (2023)
-
Structure-based insights into recognition and regulation of SAM-sensing riboswitches
Science China Life Sciences (2023)
-
SAM-VI riboswitch structure and signature for ligand discrimination
Nature Communications (2019)
-
Development of a genetically encodable FRET system using fluorescent RNA aptamers
Nature Communications (2018)
-
An excited state underlies gene regulation of a transcriptional riboswitch
Nature Chemical Biology (2017)