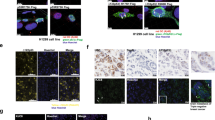
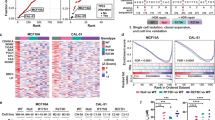
Abstract
Many p53 missense mutations possess dominant-negative activity and oncogenic gain of function. We report that for structurally destabilized p53 mutants, these effects result from mutant-induced coaggregation of wild-type p53 and its paralogs p63 and p73, thereby also inducing a heat-shock response. Aggregation of mutant p53 resulted from self-assembly of a conserved aggregation-nucleating sequence within the hydrophobic core of the DNA-binding domain, which becomes exposed after mutation. Suppressing the aggregation propensity of this sequence by mutagenesis abrogated gain of function and restored activity of wild-type p53 and its paralogs. In the p53 germline mutation database, tumors carrying aggregation-prone p53 mutations have a significantly lower frequency of wild-type allele loss as compared to tumors harboring nonaggregating mutations, suggesting a difference in clonal selection of aggregating mutants. Overall, our study reveals a novel disease mechanism for mutant p53 gain of function and suggests that, at least in some respects, cancer could be considered an aggregation-associated disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Aguzzi, A. & O'Connor, T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat. Rev. Drug Discov. 9, 237–248 (2010).
Huo, Q. Protein complexes/aggregates as potential cancer biomarkers revealed by a nanoparticle aggregation immunoassay. Colloids Surf. B Biointerfaces 78, 259–265 (2010).
Maslon, M.M. & Hupp, T.R. Drug discovery and mutant p53. Trends Cell Biol. 20, 542–555 (2010).
Olivier, M. et al. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 19, 607–614 (2002).
Ang, H.C., Joerger, A.C., Mayer, S. & Fersht, A.R. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J. Biol. Chem. 281, 21934–21941 (2006).
Gannon, J.V., Greaves, R., Iggo, R. & Lane, D.P. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 9, 1595–1602 (1990).
Joerger, A.C. & Fersht, A.R. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77, 557–582 (2008).
Chan, W.M., Siu, W.Y., Lau, A. & Poon, R.Y. How many mutant p53 molecules are needed to inactivate a tetramer? Mol. Cell. Biol. 24, 3536–3551 (2004).
Brosh, R. & Rotter, V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer 9, 701–713 (2009).
Su, X. et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467, 986–990 (2010).
Leong, C.O., Vidnovic, N., DeYoung, M.P., Sgroi, D. & Ellisen, L.W. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J. Clin. Invest. 117, 1370–1380 (2007).
Gaiddon, C., Lokshin, M., Ahn, J., Zhang, T. & Prives, C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21, 1874–1887 (2001).
Lang, G.A. et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–872 (2004).
Ostermeyer, A.G., Runko, E., Winkfield, B., Ahn, B. & Moll, U.M. Cytoplasmically sequestered wild-type p53 protein in neuroblastoma is relocated to the nucleus by a C-terminal peptide. Proc. Natl. Acad. Sci. USA 93, 15190–15194 (1996).
Johnston, J.A., Ward, C.L. & Kopito, R.R. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898 (1998).
Okorokov, A.L. & Orlova, E.V. Structural biology of the p53 tumour suppressor. Curr. Opin. Struct. Biol. 19, 197–202 (2009).
Fernandez-Escamilla, A.M., Rousseau, F., Schymkowitz, J. & Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22, 1302–1306 (2004).
Bullock, A.N. & Fersht, A.R. Rescuing the function of mutant p53. Nat. Rev. Cancer 1, 68–76 (2001).
Ishimaru, D. et al. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry 42, 9022–9027 (2003).
Kruse, J.P. & Gu, W. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. J. Biol. Chem. 284, 3250–3263 (2009).
Liang, S.H. & Clarke, M.F. A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. J. Biol. Chem. 274, 32699–32703 (1999).
Haupt, S., Berger, M., Goldberg, Z. & Haupt, Y. Apoptosis—the p53 network. J. Cell Sci. 116, 4077–4085 (2003).
Davison, T.S., Yin, P., Nie, E., Kay, C. & Arrowsmith, C.H. Characterization of the oligomerization defects of two p53 mutants found in families with Li-Fraumeni and Li-Fraumeni-like syndrome. Oncogene 17, 651–656 (1998).
Strano, S. et al. Physical and functional interaction between p53 mutants and different isoforms of p73. J. Biol. Chem. 275, 29503–29512 (2000).
Li, Y. & Prives, C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene 26, 2220–2225 (2007).
Joerger, A.C. et al. Structural evolution of p53, p63, and p73: implication for heterotetramer formation. Proc. Natl. Acad. Sci. USA 106, 17705–17710 (2009).
Rajan, R.S., Illing, M.E., Bence, N.F. & Kopito, R.R. Specificity in intracellular protein aggregation and inclusion body formation. Proc. Natl. Acad. Sci. USA 98, 13060–13065 (2001).
Cam, H. et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell 10, 281–293 (2006).
Boominathan, L. Some facts and thoughts: p73 as a tumor suppressor gene in the network of tumor suppressors. Mol. Cancer 6, 27 (2007).
Hishiya, A. & Takayama, S. Molecular chaperones as regulators of cell death. Oncogene 27, 6489–6506 (2008).
Whitesell, L. & Lindquist, S.L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5, 761–772 (2005).
Ciocca, D.R. & Calderwood, S.K. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10, 86–103 (2005).
Sedlacek, Z., Kodet, R., Poustka, A. & Goetz, P. A database of germline p53 mutations in cancer-prone families. Nucleic Acids Res. 26, 214–215 (1998).
Powell, B., Soong, R., Iacopetta, B., Seshadri, R. & Smith, D.R. Prognostic significance of mutations to different structural and functional regions of the p53 gene in breast cancer. Clin. Cancer Res. 6, 443–451 (2000).
Samowitz, W.S. et al. Prognostic significance of p53 mutations in colon cancer at the population level. Int. J. Cancer 99, 597–602 (2002).
Davison, T.S. et al. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J. Biol. Chem. 274, 18709–18714 (1999).
Finlay, C.A. et al. Activating mutations for transformation by p53 produce a gene product that forms an hsc70-p53 complex with an altered half-life. Mol. Cell. Biol. 8, 531–539 (1988).
Milner, J. & Medcalf, E.A. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell 65, 765–774 (1991).
Friedman, P.N., Chen, X., Bargonetti, J. & Prives, C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc. Natl. Acad. Sci. USA 90, 3319–3323 (1993).
Goh, A.M., Coffill, C.R. & Lane, D.P. The role of mutant p53 in human cancer. J. Pathol. 223, 116–126 (2011).
Flores, E.R. et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7, 363–373 (2005).
Bensaad, K. et al. Change of conformation of the DNA-binding domain of p53 is the only key element for binding of and interference with p73. J. Biol. Chem. 278, 10546–10555 (2003).
Bullock, A.N. et al. Thermodynamic stability of wild-type and mutant p53 core domain. Proc. Natl. Acad. Sci. USA 94, 14338–14342 (1997).
Rotter, V. p53, a transformation-related cellular-encoded protein, can be used as a biochemical marker for the detection of primary mouse tumor cells. Proc. Natl. Acad. Sci. USA 80, 2613–2617 (1983).
Moll, U.M., Riou, G. & Levine, A.J. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc. Natl. Acad. Sci. USA 89, 7262–7266 (1992).
Ostermeyer, A.G., Runko, E., Winkfield, B., Ahn, B. & Moll, U.M. Cytoplasmically sequestered wild-type p53 protein in neuroblastoma is relocated to the nucleus by a C-terminal peptide. Proc. Natl. Acad. Sci. USA 93, 15190–15194 (1996).
Fernandez-Escamilla, A.M., Rousseau, F., Schymkowitz, J. & Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22, 1302–1306 (2004).
Bullock, A.N., Henckel, J. & Fersht, A.R. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene 19, 1245–1256 (2000).
Schymkowitz, J. et al. The FoldX web server: an online force field. Nucleic Acids Res. 33, W382–W388 (2005).
Di Como, C.J., Gaiddon, C. & Prives, C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 19, 1438–1449 (1999).
Acknowledgements
The VIB Switch Laboratory was supported by the Research Foundation Flanders and the Agency for Innovation by Science and Technology Flanders. J.X. and A.Z. were supported by Linking Sino-European Universities through Mobility and National Natural Science Foundation of China (81000861) and the Research Foundation Flanders, respectively. D.L. was supported by the Research Council Katholieke Universiteit Leuven, Center of Excellence (KUL PFV/10/016 SymBioSys) and the Stichting Tegen Kanker. We thank G. Peuteman, D. Smeets and T. Van Brussel for technical assistance. We thank G. Lozano for access to tumor tissues from transgenic mice and G. Melino for plasmids.
Author information
Authors and Affiliations
Contributions
J.X. performed BN-PAGE, immunofluorescence, immunoprecipitation, qPCR and analyzed clinical data; J. Reumers analyzed stability of p53 mutants by FoldX; J.R.C. studied peptide aggregation and analyzed tumor tissues with J.X.; R.G. purified p53 and performed electron microscopy and FTIR; J. Rozenski performed ESI-MS study; A.C. provided clinical tissue samples; F.D.S., S.R., A.Z. and J.-C.M. did cellular experiments; D.L. sequenced TP53 in tumors; Y.-A.S. prepared tissue sections from mice; J.X., F.R., J.S. and F.D.S. wrote the manuscript; F.R. and J.S. formulated the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Figures 1–18 and Supplementary Table 1 (PDF 8472 kb)
Rights and permissions
About this article
Cite this article
Xu, J., Reumers, J., Couceiro, J. et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol 7, 285–295 (2011). https://doi.org/10.1038/nchembio.546
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.546
This article is cited by
-
Amyloid aggregates induced by the p53-R280T mutation lead to loss of p53 function in nasopharyngeal carcinoma
Cell Death & Disease (2024)
-
Phase separations in oncogenesis, tumor progressions and metastasis: a glance from hallmarks of cancer
Journal of Hematology & Oncology (2023)
-
Amyloid-like p53 as prognostic biomarker in serous ovarian cancer—a study of the OVCAD consortium
Oncogene (2023)
-
Drugging p53 in cancer: one protein, many targets
Nature Reviews Drug Discovery (2023)
-
Actionable cancer vulnerability due to translational arrest, p53 aggregation and ribosome biogenesis stress evoked by the disulfiram metabolite CuET
Cell Death & Differentiation (2023)