Abstract
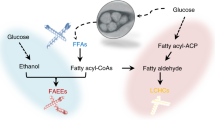
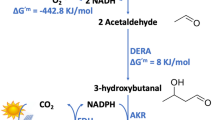
Living systems have evolved remarkable molecular functions that can be redesigned for in vivo chemical synthesis as we gain a deeper understanding of the underlying biochemical principles for de novo construction of synthetic pathways. We have focused on developing pathways for next-generation biofuels as they require carbon to be channeled to product at quantitative yields. However, these fatty acid–inspired pathways must manage the highly reversible nature of the enzyme components. For targets in the biodiesel range, the equilibrium can be driven to completion by physical sequestration of an insoluble product, which is a mechanism unavailable to soluble gasoline-sized products. In this work, we report the construction of a chimeric pathway assembled from three different organisms for the high-level production of n-butanol (4,650 ± 720 mg l−1) that uses an enzymatic chemical reaction mechanism in place of a physical step as a kinetic control element to achieve high yields from glucose (28%).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Stephanopoulos, G. & Sinskey, A.J. Metabolic engineering—methodologies and future prospects. Trends Biotechnol. 11, 392–396 (1993).
Rohlin, L., Oh, M.K. & Liao, J.C. Microbial pathway engineering for industrial processes: Evolution, combinatorial biosynthesis and rational design. Curr. Opin. Microbiol. 4, 330–335 (2001).
Keasling, J.D. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 3, 64–76 (2008).
Ro, D.K. et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943 (2006).
Draths, K.M., Knop, D.R. & Frost, J.W. Shikimic acid and quinic acid: Replacing isolation from plant sources with recombinant microbial biocatalysis. J. Am. Chem. Soc. 121, 1603–1604 (1999).
Biebl, H., Menzel, K., Zeng, A.P. & Deckwer, W.D. Microbial production of 1,3-propanediol. Appl. Microbiol. Biotechnol. 52, 289–297 (1999).
Bragg, J.R., Prince, R.C., Harner, E.J. & Ronald, M.A. Effectiveness of bioremediation for the Exxon Valdez oil spill. Nature 368, 413–418 (1994).
Fischer, C.R., Klein-Marcuschamer, D. & Stephanopoulos, G. Selection and optimization of microbial hosts for biofuels production. Metab. Eng. 10, 295–304 (2008).
Atsumi, S. & Liao, J.C. Metabolic engineering for advanced biofuels production from Escherichia coli. Curr. Opin. Biotechnol. 19, 414–419 (2008).
Atsumi, S., Hanai, T. & Liao, J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89 (2008).
Steen, E.J. et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562 (2010).
Schirmer, A., Rude, M.A., Li, X., Popova, E. & del Cardayre, S.B. Microbial biosynthesis of alkanes. Science 329, 559–562 (2010).
Sheehan, J., Dunahay, T., Benemann, J. & Roessler, P. A look back at the U.S. Department of Energy's Aquatic Species Program: Biodiesel from algae (U.S. Department of Energy, Office of Fuels Development NREL/TP-580–24190), National Renewable Energy Laboratory (1998).
Zaslavskaia, L.A. et al. Trophic conversion of an obligate photoautotrophic organism through metabolic engineering. Science 292, 2073–2075 (2001).
Dürre, P. Biobutanol: An attractive biofuel. Biotechnol. J. 2, 1525–1534 (2007).
Lee, S.Y. et al. Fermentative butanol production by clostridia. Biotechnol. Bioeng. 101, 209–228 (2008).
Papoutsakis, E.T. Engineering solventogenic clostridia. Curr. Opin. Biotechnol. 19, 420–429 (2008).
Nair, R.V., Green, E.M., Watson, D.E., Bennett, G.N. & Papoutsakis, E.T. Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J. Bacteriol. 181, 319–330 (1999).
Scotcher, M.C., Rudolph, F.B. & Bennett, G.N. Expression of abrB310 and sinR, and effects of decreased abrB310 expression on the transition from acidogenesis to solventogenesis, in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 71, 1987–1995 (2005).
Atsumi, S. et al. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10, 305–311 (2008).
Steen, E.J. et al. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb. Cell Fact. 7, 36 (2008).
Nielsen, D.R. et al. Engineering alternative butanol production platforms in heterologous bacteria. Metab. Eng. 11, 262–273 (2009).
Inui, M. et al. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl. Microbiol. Biotechnol. 77, 1305–1316 (2008).
Peoples, O.P. & Sinskey, A.J. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J. Biol. Chem. 264, 15298–15303 (1989).
Lynen, F. & Ochoa, S. Enzymes of fatty acid metabolism. Biochim. Biophys. Acta 12, 299–314 (1953).
Leaf, T.A. & Srienc, F. Metabolic modeling of polyhydroxybutyrate biosynthesis. Biotechnol. Bioeng. 57, 557–570 (1998).
Waterson, R.M., Castellino, F.J., Hass, G.M. & Hill, R.L. Purification and characterization of crotonase from Clostridium acetobutylicum. J. Biol. Chem. 247, 5266–5271 (1972).
Boynton, Z.L., Bennet, G.N. & Rudolph, F.B. Cloning, sequencing, and expression of clustered genes encoding β-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 178, 3015–3024 (1996).
Wallace, K.K. et al. Purification of crotonyl-CoA reductase from Streptomyces collinus and cloning, sequencing and expression of the corresponding gene in Escherichia coli. Eur. J. Biochem. 233, 954–962 (1995).
Fontaine, L. et al. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184, 821–830 (2002).
Slater, S.C., Voige, W.H. & Dennis, D.E. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-β-hydroxybutyrate biosynthetic pathway. J. Bacteriol. 170, 4431–4436 (1988).
Li, F. et al. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 190, 843–850 (2008).
Hoffmeister, M., Piotrowski, M., Nowitzki, U. & Martin, W. Mitochondrial trans-2-enoyl-CoA reductase of wax ester fermentation from Euglena gracilis defines a new family of enzymes involved in lipid synthesis. J. Biol. Chem. 280, 4329–4338 (2005).
Tucci, S. & Martin, W. A novel prokaryotic trans-2-enoyl-CoA reductase from the spirochete Treponema denticola. FEBS Lett. 581, 1561–1566 (2007).
Bennett, B.D. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009).
Erb, T.J., Brecht, V., Fuchs, G., Muller, M. & Alber, B.E. Carboxylation mechanism and stereochemistry of crotonyl-CoA carboxylase/reductase, a carboxylating enoyl-thioester reductase. Proc. Natl. Acad. Sci. USA 106, 8871–8876 (2009).
Wakil, S.J. Studies on the fatty acid oxidizing system of animal tissues. IX. Stereospecificity of unsaturated acyl CoA hydrase. Biochim. Biophys. Acta 19, 497–504 (1956).
Roca, C., Nielsen, J. & Olsson, L. Metabolic engineering of ammonium assimilation in xylose-fermenting Saccharomyces cerevisiae improves ethanol production. Appl. Environ. Microbiol. 69, 4732–4736 (2003).
Sanchez, A.M., Andrews, J., Hussein, I., Bennett, G.N. & San, K.Y. Effect of overexpression of a soluble pyridine nucleotide transhydrogenase (UdhA) on the production of poly(3-hydroxybutyrate) in Escherichia coli. Biotechnol. Prog. 22, 420–425 (2006).
Fukui, T., Shiomi, N. & Doi, Y. Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J. Bacteriol. 180, 667–673 (1998).
Agius, L. & Seratt, H.S.A. Channeling in intermediary metabolism. (Portland Press, Ltd., London, 1996).
Hopwood, D.A. & Sherman, D.H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 24, 37–66 (1990).
White, S.W., Zheng, J., Zhang, Y.M. & Rock, C.O. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831 (2005).
Kunau, W.H., Dommes, V. & Schulz, H. β-oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: A century of continued progress. Prog. Lipid Res. 34, 267–342 (1995).
An, S., Kumar, R., Sheets, E.D. & Benkovic, S.J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320, 103–106 (2008).
Dueber, J.E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759 (2009).
Yeates, T.O., Kerfeld, C.A., Heinhorst, S., Cannon, G.C. & Shively, J.M. Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6, 681–691 (2008).
Wu, W.-J. et al. Stereospecificity of the reaction catalyzed by enoyl-CoA hydratase. J. Am. Chem. Soc. 122, 3987–3994 (2000).
Wakil, S.J., Green, D.E., Mii, S. & Mahler, H.R. Studies on the fatty acid oxidizing system of animal tissues. VI. β-Hydroxyacyl coenzyme A dehydrogenase. J. Biol. Chem. 207, 631–638 (1954).
Tummala, S.B., Welker, N.E. & Papoutsakis, E.T. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J. Bacteriol. 185, 1923–1934 (2003).
Acknowledgements
We thank K. Hirano for her work on the ter gene assembly during her rotation. B.B.B.-W. would like to thank the Aldo DeBenedictis Fund for a predoctoral fellowship, and R.J.B. would like to acknowledge the University of California, Berkeley, Summer Undergraduate Research Fellowship program. This work was funded by generous support from University of California, Berkeley, the Camille and Henry Dreyfus Foundation, the Arnold and Mabel Beckman Foundation and the Dow Sustainable Products and Solutions Program.
Author information
Authors and Affiliations
Contributions
R.J.B. constructed the plasmids for Ccr quantification and measured n-butanol production and Ccr-Stag levels in these strains. B.B.B.-W. carried out the remaining experiments. B.B.B.-W and M.C.Y.C. planned the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Figures 1–11 and Supplementary Tables 1–8 (PDF 1685 kb)
Rights and permissions
About this article
Cite this article
Bond-Watts, B., Bellerose, R. & Chang, M. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol 7, 222–227 (2011). https://doi.org/10.1038/nchembio.537
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.537
This article is cited by
-
Unlocking the potentials of cyanobacterial photosynthesis for directly converting carbon dioxide into glucose
Nature Communications (2023)
-
Microbial Polyhydroxyalkanoates (PHAs): A Review on Biosynthesis, Properties, Fermentation Strategies and Its Prospective Applications for Sustainable Future
Journal of Polymers and the Environment (2022)
-
Microbial production of advanced biofuels
Nature Reviews Microbiology (2021)
-
Two steps to sustainable polymers
Nature Chemistry (2021)
-
A dual cellular–heterogeneous catalyst strategy for the production of olefins from glucose
Nature Chemistry (2021)