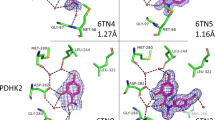
Abstract
Fragment screens have successfully identified new scaffolds in drug discovery, often with relatively high hit rates (5%) using small screening libraries (1,000–10,000 compounds). This raises two questions: would other noteworthy chemotypes be found were one to screen all commercially available fragments (>300,000), and does the success rate imply low specificity of fragments? We used molecular docking to screen large libraries of fragments against CTX-M β-lactamase. We identified ten millimolar-range inhibitors from the 69 compounds tested. The docking poses corresponded closely to the crystallographic structures subsequently determined. Notably, these initial low-affinity hits showed little specificity between CTX-M and an unrelated β-lactamase, AmpC, which is unusual among β-lactamase inhibitors. This is consistent with the idea that the high hit rates among fragments correlate to a low initial specificity. As the inhibitors were progressed, both specificity and affinity rose together, yielding to our knowledge the first micromolar-range noncovalent inhibitors against a class A β-lactamase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Rees, D.C., Congreve, M., Murray, C.W. & Carr, R. Fragment-based lead discovery. Nat. Rev. Drug Discov. 3, 660–672 (2004).
Congreve, M., Chessari, G., Tisi, D. & Woodhead, A.J. Recent developments in fragment-based drug discovery. J. Med. Chem. 51, 3661–3680 (2008).
Murray, C.W. et al. Application of fragment screening by X-ray crystallography to beta-secretase. J. Med. Chem. 50, 1116–1123 (2007).
Haydon, D.J. et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321, 1673–1675 (2008).
Card, G.L. et al. A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design. Nat. Biotechnol. 23, 201–207 (2005).
Edwards, P.D. et al. Application of fragment-based lead generation to the discovery of novel, cyclic amidine beta-secretase inhibitors with nanomolar potency, cellular activity, and high ligand efficiency. J. Med. Chem. 50, 5912–5925 (2007).
Fahr, B.T. et al. Tethering identifies fragment that yields potent inhibitors of human caspase-1. Bioorg. Med. Chem. Lett. 16, 559–562 (2006).
Oltersdorf, T. et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681 (2005).
Shuker, S.B., Hajduk, P.J., Meadows, R.P. & Fesik, S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274, 1531–1534 (1996).
Pellecchia, M. et al. Perspectives on NMR in drug discovery: a technique comes of age. Nat. Rev. Drug Discov. 7, 738–745 (2008).
Hartshorn, M.J. et al. Fragment-based lead discovery using X-ray crystallography. J. Med. Chem. 48, 403–413 (2005).
Sweeney, Z.K. et al. Discovery of triazolinone non-nucleoside inhibitors of HIV reverse transcriptase. Bioorg. Med. Chem. Lett. 18, 4348–4351 (2008).
Huang, J.W. et al. Fragment-based design of small molecule X-linked inhibitor of apoptosis protein inhibitors. J. Med. Chem. 51, 7111–7118 (2008).
Marcou, G. & Rognan, D. Optimizing fragment and scaffold docking by use of molecular interaction fingerprints. J. Chem. Inf. Model. 47, 195–207 (2007).
Hubbard, R.E., Chen, I. & Davis, B. Informatics and modeling challenges in fragment-based drug discovery. Curr. Opin. Drug Discov. Devel. 10, 289–297 (2007).
Klebe, G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov. Today 11, 580–594 (2006).
Hann, M.M., Leach, A.R. & Harper, G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J. Chem. Inf. Comput. Sci. 41, 856–864 (2001).
Bonnet, R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48, 1–14 (2004).
Chen, Y., Delmas, J., Sirot, J., Shoichet, B. & Bonnet, R. Atomic resolution structures of CTX-M beta-lactamases: extended spectrum activities from increased mobility and decreased stability. J. Mol. Biol. 348, 349–362 (2005).
Oprea, T.I., Davis, A.M., Teague, S.J. & Leeson, P.D. Is there a difference between leads and drugs? A historical perspective. J. Chem. Inf. Comput. Sci. 41, 1308–1315 (2001).
Massova, I. & Mobashery, S. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob. Agents Chemother. 42, 1–17 (1998).
Payne, D.J., Gwynn, M.N., Holmes, D.J. & Pompliano, D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 (2007).
O'Shea, R. & Moser, H.E. Physicochemical properties of antibacterial compounds: implications for drug discovery. J. Med. Chem. 51, 2871–2878 (2008).
Beadle, B.M., Trehan, I., Focia, P. & Shoichet, B.K. Structural milestones in the pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC b-lactamase. Structure 10, 413–424 (2002).
Kumar, S., Pearson, A.L. & Pratt, R.F. Design, synthesis, and evaluation of alpha-ketoheterocycles as class C beta-lactamase inhibitors. Bioorg. Med. Chem. 9, 2035–2044 (2001).
Ibuka, A.S. et al. Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for catalytic reaction and substrate specificity expansion. Biochemistry 42, 10634–10643 (2003).
Chen, Y., Shoichet, B. & Bonnet, R. Structure, function, and inhibition along the reaction coordinate of CTX-M beta-lactamases. J. Am. Chem. Soc. 127, 5423–5434 (2005).
Babaoglu, K. et al. Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase. J. Med. Chem. 51, 2502–2511 (2008).
Powers, R.A., Morandi, F. & Shoichet, B.K. Structure-based discovery of a novel, noncovalent inhibitor of AmpC beta-lactamase. Structure 10, 1013–1023 (2002).
Chen, Y., Bonnet, R. & Shoichet, B.K. The acylation mechanism of CTX-M beta-lactamase at 0.88 a resolution. J. Am. Chem. Soc. 129, 5378–5380 (2007).
Graves, A.P. et al. Rescoring docking hit lists for model cavity sites: predictions and experimental testing. J. Mol. Biol. 377, 914–934 (2008).
Lorber, D.M. & Shoichet, B.K. Hierarchical docking of databases of multiple ligand conformations. Curr. Top. Med. Chem. 5, 739–749 (2005).
Irwin, J.J. & Shoichet, B.K. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 (2005).
Lorber, D.M. & Shoichet, B.K. Flexible ligand docking using conformational ensembles. Protein Sci. 7, 938–950 (1998).
Huang, N., Shoichet, B.K. & Irwin, J.J. Benchmarking sets for molecular docking. J. Med. Chem. 49, 6789–6801 (2006).
Meng, E.C., Gschwend, D.C., Blaney, J.M. & Kuntz, I.D. Orientational sampling and rigid-body minimization in molecular docking. Proteins 17, 266–278 (1993).
Shoichet, B.K., Leach, A.R. & Kuntz, I.D. Ligand solvation in molecular docking. Proteins 34, 4–16 (1999).
McGovern, S.L., Caselli, E., Grigorieff, N. & Shoichet, B.K. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 45, 1712–1722 (2002).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50, 760–763 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
This work was supported by US National Institutes of Health grants GM63813 and GM59957 (to B.K.S.). We thank D.G. Teotico (University of California, San Francisco) for providing the AmpC inhibitors and for insightful discussions, and J. Hert and C. Laggner for assistance with similarity search. We thank D.G. Teotico, S. Boyce, M. Mysinger and J. Irwin for reading the manuscript. We also thank R. Bonnet.
Author information
Authors and Affiliations
Contributions
This project was designed by Y.C. and B.K.S. together; all the experiments were undertaken by Y.C. Both authors contributed to interpreting the results and writing the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Tables 1 and 2 (PDF 271 kb)
Rights and permissions
About this article
Cite this article
Chen, Y., Shoichet, B. Molecular docking and ligand specificity in fragment-based inhibitor discovery. Nat Chem Biol 5, 358–364 (2009). https://doi.org/10.1038/nchembio.155
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.155
This article is cited by
-
Synthon-based ligand discovery in virtual libraries of over 11 billion compounds
Nature (2022)
-
ASFP (Artificial Intelligence based Scoring Function Platform): a web server for the development of customized scoring functions
Journal of Cheminformatics (2021)
-
A practical guide to large-scale docking
Nature Protocols (2021)
-
Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site
Nature Communications (2020)
-
Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography
Scientific Reports (2019)