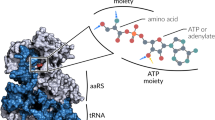
Abstract
At the time of its discovery four decades ago, the genetic code was viewed as the result of a “frozen accident.” Our current knowledge of the translation process and of the detailed structure of its components highlights the roles of RNA structure (in mRNA and tRNA), RNA modification (in tRNA), and aminoacyl-tRNA synthetase diversity in the evolution of the genetic code. The diverse assortment of codon reassignments present in subcellular organelles and organisms of distinct lineages has 'thawed' the concept of a universal immutable code; it may not be accidental that out of more than 140 amino acids found in natural proteins, only two (selenocysteine and pyrrolysine) are known to have been added to the standard 20-member amino acid alphabet. The existence of phosphoseryl-tRNA (in the form of tRNACys and tRNASec) may presage the discovery of other cotranslationally inserted modified amino acids.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Ibba, M. & Söll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 (2000).
Ibba, M. & Söll, D. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev. 18, 731–738 (2004).
Uy, R. & Wold, F. Posttranslational covalent modification of proteins. Science 198, 890–896 (1977).
Furter, R. Expansion of the genetic code: site-directed p-fluoro-phenylalanine incorporation in Escherichia coli. Protein Sci. 7, 419–426 (1998).
Wang, L., Xie, J. & Schultz, P.G. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 35, 225–249 (2006).
Köhrer, C. & RajBhandary, U.L. in The Aminoacyl-tRNA Synthetases (eds. Ibba, M., Francklyn, C.S. & Cusack, S.) 353–363 (Landes Bioscience, Georgetown, Texas, USA, 2005).
Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 38, 367–379 (1968).
Woese, C.R. On the evolution of the genetic code. Proc. Natl. Acad. Sci. USA 54, 1546–1552 (1965).
Osawa, S., Jukes, T.H., Watanabe, K. & Muto, A. Recent evidence for evolution of the genetic code. Microbiol. Rev. 56, 229–264 (1992).
Knight, R.D., Freeland, S.J. & Landweber, L.F. Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 2, 49–58 (2001).
Santos, M.A., Moura, G., Massey, S.E. & Tuite, M.F. Driving change: the evolution of alternative genetic codes. Trends Genet. 20, 95–102 (2004).
Miranda, I., Silva, R. & Santos, M.A. Evolution of the genetic code in yeasts. Yeast 23, 203–213 (2006).
Crick, F.H.C. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19, 548–555 (1966).
Söll, D. et al. Specificity of sRNA for recognition of codons as studied by the ribosomal binding technique. J. Mol. Biol. 19, 556–573 (1966).
Kisselev, L., Ehrenberg, M. & Frolova, L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 22, 175–182 (2003).
Eggertsson, G. & Söll, D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol. Rev. 52, 354–374 (1988).
Söll, D. Genetic code: enter a new amino acid. Nature 331, 662–663 (1988).
Schön, A., Böck, A., Ott, G., Sprinzl, M. & Söll, D. The selenocysteine-inserting opal suppressor serine tRNA from E. coli is highly unusual in structure and modification. Nucleic Acids Res. 17, 7159–7165 (1989).
Böck, A., Thanbichler, M., Rother, M. & Resch, A. in The Aminoacyl-tRNA Synthetases (eds. Ibba, M., Francklyn, C.S. & Cusack, S.) 320–327 (Landes Bioscience, Georgetown, Texas, USA, 2005).
Hao, B. et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 296, 1462–1466 (2002).
Srinivasan, G., James, C.M. & Krzycki, J.A. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 296, 1459–1462 (2002).
Polycarpo, C. et al. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. USA 101, 12450–12454 (2004).
Blight, S.K. et al. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature 431, 333–335 (2004).
Heckman, J.E., Sarnoff, J., Alzner-DeWeerd, B., Yin, S. & RajBhandary, U.L. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc. Natl. Acad. Sci. USA 77, 3159–3163 (1980).
Yarus, M. Translational efficiency of transfer RNA's: uses of an extended anticodon. Science 218, 646–652 (1982).
Agris, P.F. Decoding the genome: a modified view. Nucleic Acids Res. 32, 223–238 (2004).
Tomita, K., Ueda, T. & Watanabe, K. 7-Methylguanosine at the anticodon wobble position of squid mitochondrial tRNASer GCU: molecular basis for assignment of AGA/AGG codons as serine in invertebrate mitochondria. Biochim. Biophys. Acta 1399, 78–82 (1998).
Tomita, K., Ueda, T. & Watanabe, K. The presence of pseudouridine in the anticodon alters the genetic code: a possible mechanism for assignment of the AAA lysine codon as asparagine in echinoderm mitochondria. Nucleic Acids Res. 27, 1683–1689 (1999).
Suzuki, T., Ueda, T. & Watanabe, K. The 'polysemous' codon–a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 16, 1122–1134 (1997).
Min, B. et al. Protein synthesis in Escherichia coli with mischarged tRNA. J. Bacteriol. 185, 3524–3526 (2003).
Mehl, R.A. et al. Generation of a bacterium with a 21 amino acid genetic code. J. Am. Chem. Soc. 125, 935–939 (2003).
Vetsigian, K., Woese, C. & Goldenfeld, N. Collective evolution and the genetic code. Proc. Natl. Acad. Sci. USA 103, 10696–10701 (2006).
Rayman, M.P. The importance of selenium to human health. Lancet 356, 233–241 (2000).
Cone, J.E., Del Rio, R.M., Davis, J.N. & Stadtman, T.C. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc. Natl. Acad. Sci. USA 73, 2659–2663 (1976).
Fu, L.H. et al. A selenoprotein in the plant kingdom. Mass spectrometry confirms that an opal codon (UGA) encodes selenocysteine in Chlamydomonas reinhardtii glutathione peroxidase. J. Biol. Chem. 277, 25983–25991 (2002).
Obata, T. & Shiraiwa, Y. A novel eukaryotic selenoprotein in the haptophyte alga Emiliania huxleyi. J. Biol. Chem. 280, 18462–18468 (2005).
Hatfield, D., Choi, I.S., Mischke, S. & Owens, L.D. Selenocysteyl-tRNAs recognize UGA in Beta vulgaris, a higher plant, and in Gliocladium virens, a filamentous fungus. Biochem. Biophys. Res. Commun. 184, 254–259 (1992).
Kryukov, G.V. et al. Characterization of mammalian selenoproteomes. Science 300, 1439–1443 (2003).
Kryukov, G.V. & Gladyshev, V.N. The prokaryotic selenoproteome. EMBO Rep. 5, 538–543 (2004).
Johansson, L., Gafvelin, G. & Arner, E.S. Selenocysteine in proteins-properties and biotechnological use. Biochim. Biophys. Acta 1726, 1–13 (2005).
Zhang, Y., Romero, H., Salinas, G. & Gladyshev, V.N. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox-active cysteine residues. Genome Biol. 7, R94 (2006).
Baron, C., Heider, J. & Böck, A. Mutagenesis of selC, the gene for the selenocysteine-inserting tRNA-species in E. coli: effects on in vivo function. Nucleic Acids Res. 18, 6761–6766 (1990).
Forchhammer, K., Boesmiller, K. & Böck, A. The function of selenocysteine synthase and SELB in the synthesis and incorporation of selenocysteine. Biochimie 73, 1481–1486 (1991).
Tormay, P. et al. Bacterial selenocysteine synthase–structural and functional properties. Eur. J. Biochem. 254, 655–661 (1998).
Leinfelder, W., Stadtman, T.C. & Böck, A. Occurrence in vivo of selenocysteyl-tRNASerUCA in Escherichia coli. Effect of sel mutations. J. Biol. Chem. 264, 9720–9723 (1989).
Forster, C., Ott, G., Forchhammer, K. & Sprinzl, M. Interaction of a selenocysteine-incorporating tRNA with elongation factor Tu from E. coli. Nucleic Acids Res. 18, 487–491 (1990).
Forchhammer, K., Leinfelder, W. & Böck, A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature 342, 453–456 (1989).
Wu, X.Q. & Gross, H.J. The long extra arms of human tRNA(Ser)Sec and tRNASer function as major identify elements for serylation in an orientation-dependent, but not sequence-specific manner. Nucleic Acids Res. 21, 5589–5594 (1993).
Sturchler-Pierrat, C. et al. Selenocysteylation in eukaryotes necessitates the uniquely long aminoacyl acceptor stem of selenocysteine tRNASec. J. Biol. Chem. 270, 18570–18574 (1995).
Ohama, T., Yang, D.C. & Hatfield, D.L. Selenocysteine tRNA and serine tRNA are aminoacylated by the same synthetase, but may manifest different identities with respect to the long extra arm. Arch. Biochem. Biophys. 315, 293–301 (1994).
Geslain, R. et al. Trypanosoma seryl-tRNA synthetase is a metazoan-like enzyme with high affinity for tRNASec. J. Biol. Chem., published online 13 October 2006 (doi:10.1074/jbc.M607862200).
Kaiser, J.T. et al. Structural and functional investigation of a putative archaeal selenocysteine synthase. Biochemistry 44, 13315–13327 (2005).
Rother, M., Wilting, R., Commans, S. & Böck, A. Identification and characterization of the selenocysteine-specific translation factor SelB from the archaeon Methanococcus jannaschii. J. Mol. Biol. 299, 351–358 (2000).
Bilokapic, S., Korencic, D., Söll, D. & Weygand-Durasevic, I. The unusual methanogenic seryl-tRNA synthetase recognizes tRNASer species from all three kingdoms of life. Eur. J. Biochem. 271, 694–702 (2004).
Allmang, C. & Krol, A. Selenoprotein synthesis: UGA does not end the story. Biochimie 88, 1561–1571 (2006).
Carlson, B.A. et al. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl. Acad. Sci. USA 101, 12848–12853 (2004).
Mäenpää, P.H. & Bernfield, M.R. A specific hepatic transfer RNA for phosphoserine. Proc. Natl. Acad. Sci. USA 67, 688–695 (1970).
Sharp, S.J. & Stewart, T.S. The characterization of phosphoseryl tRNA from lactating bovine mammary gland. Nucleic Acids Res. 4, 2123–2136 (1977).
Sauerwald, A. et al. RNA-dependent cysteine biosynthesis in archaea. Science 307, 1969–1972 (2005).
Gelpi, C., Sontheimer, E.J. & Rodriguez-Sanchez, J.L. Autoantibodies against a serine tRNA-protein complex implicated in cotranslational selenocysteine insertion. Proc. Natl. Acad. Sci. USA 89, 9739–9743 (1992).
Kernebeck, T., Lohse, A.W. & Grötzinger, J. A bioinformatical approach suggests the function of the autoimmune hepatitis target antigen soluble liver antigen/liver pancreas. Hepatology 34, 230–233 (2001).
Yuan, J. et al. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl. Acad. Sci. USA 103, 18923–18927 (2006).
Small-Howard, A. et al. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol. Cell. Biol. 26, 2337–2346 (2006).
Guimaraes, M.J. et al. Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: is there an autoregulatory mechanism in selenocysteine metabolism? Proc. Natl. Acad. Sci. USA 93, 15086–15091 (1996).
Boone, D.R., Whitman, W.B. & Rouvière, P. in Methanogenesis (ed. Ferry, J.G.) 35–80 (Chapman & Hall, New York, 1993).
Krzycki, J.A. Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 8, 484–491 (2004).
Hao, B. et al. Reactivity and chemical synthesis of L-pyrrolysine- the 22nd genetically encoded amino acid. Chem. Biol. 11, 1317–1324 (2004).
Soares, J.A. et al. The residue mass of L-pyrrolysine in three distinct methylamine methyltransferases. J. Biol. Chem. 280, 36962–36969 (2005).
Polycarpo, C. et al. Activation of the pyrrolysine suppressor tRNA requires formation of a ternary complex with class I and class II lysyl-tRNA synthetases. Mol. Cell 12, 287–294 (2003).
Zhang, Y., Baranov, P.V., Atkins, J.F. & Gladyshev, V.N. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J. Biol. Chem. 280, 20740–20751 (2005).
Fu, S.L. & Dean, R.T. Structural characterization of the products of hydroxyl-radical damage to leucine and their detection on proteins. Biochem. J. 324, 41–48 (1997).
Zhang, M. et al. Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry 43, 12539–12548 (2004).
Théobald-Dietrich, A., Frugier, M., Giegé, R. & Rudinger-Thirion, J. Atypical archaeal tRNA pyrrolysine transcript behaves towards EF-Tu as a typical elongator tRNA. Nucleic Acids Res. 32, 1091–1096 (2004).
Namy, O., Rousset, J.P., Napthine, S. & Brierley, I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell 13, 157–168 (2004).
Théobald-Dietrich, A., Giegé, R. & Rudinger-Thirion, J. Evidence for the existence in mRNAs of a hairpin element responsible for ribosome dependent pyrrolysine insertion into proteins. Biochimie 87, 813–817 (2005).
Polycarpo, C.R. et al. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett., published online 20 November 2006 (doi:10.1016/j.febslet.2006.11.028).
Li, T. et al. Cysteinyl-tRNA formation: the last puzzle of aminoacyl-tRNA synthesis. FEBS Lett. 462, 302–306 (1999).
Stathopoulos, C. et al. Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis. Proc. Natl. Acad. Sci. USA 98, 14292–14297 (2001).
Hohn, M.J., Park, H.-S., O'Donoghue, P., Schnitzbauer, M. & Söll, D. Emergence of the universal genetic code imprinted in an RNA record. Proc. Natl. Acad. Sci. USA 103, 18095–18100 (2006).
Ibba, M., Bono, J.L., Rosa, P.A. & Söll, D. Archaeal-type lysyl-tRNA synthetase in the Lyme disease spirochete Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 94, 14383–14388 (1997).
Korencic, D., Polycarpo, C., Weygand-Durasevic, I. & Söll, D. Differential modes of transfer RNASer recognition in Methanosarcina barkeri. J. Biol. Chem. 279, 48780–48786 (2004).
Mazauric, M.H. et al. Glycyl-tRNA synthetase from Thermus thermophilus—wide structural divergence with other prokaryotic glycyl-tRNA synthetases and functional inter-relation with prokaryotic and eukaryotic glycylation systems. Eur. J. Biochem. 251, 744–757 (1998).
Mazauric, M.H., Roy, H. & Kern, D. tRNA glycylation system from Thermus thermophilus. tRNAGly identity and functional interrelation with the glycylation systems from other phylae. Biochemistry 38, 13094–13105 (1999).
Murphy, F.V. IV, Ramakrishnan, V., Malkiewicz, A. & Agris, P.F. The role of modifications in codon discrimination by tRNALys UUU . Nat. Struct. Mol. Biol. 11, 1186–1191 (2004).
Acknowledgements
We thank P. Agris for critical comments on the paper. S.P. holds a fellowship of the Yale University School of Medicine MD/PhD Program. Work in the authors' laboratory was supported by grants from the US National Institute of General Medical Sciences (GM22854), the US Department of Energy (DE-FG02-98ER20311) and the US National Science Foundation (DBI-0535566).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ambrogelly, A., Palioura, S. & Söll, D. Natural expansion of the genetic code. Nat Chem Biol 3, 29–35 (2007). https://doi.org/10.1038/nchembio847
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio847
This article is cited by
-
Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-α-amino acids in vitro and in vivo
Nature Chemistry (2023)
-
Cell-free Biosynthesis of Peptidomimetics
Biotechnology and Bioprocess Engineering (2023)
-
Deciphering the conformational landscape of few selected aromatic noncoded amino acids (NCAAs) for applications in rational design of peptide therapeutics
Amino Acids (2022)
-
Feeding intact proteins, peptides, or free amino acids to monogastric farm animals
Amino Acids (2022)
-
Review: microbial transformations of human bile acids
Microbiome (2021)