Abstract
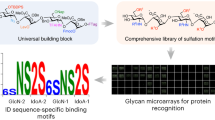
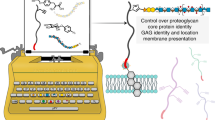
Although glycosaminoglycans contribute to diverse physiological processes1,2,3,4, an understanding of their molecular mechanisms has been hampered by the inability to access homogeneous glycosaminoglycan structures. Here, we assembled well-defined chondroitin sulfate oligosaccharides using a convergent, synthetic approach that permits installation of sulfate groups at precise positions along the carbohydrate backbone. Using these defined structures, we demonstrate that specific sulfation motifs function as molecular recognition elements for growth factors and modulate neuronal growth. These results provide both fundamental insights into the role of sulfation and direct evidence for a 'sulfation code' whereby glycosaminoglycans encode functional information in a sequence-specific manner analogous to that of DNA, RNA and proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hacker, U., Nybakken, K. & Perrimon, N. Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol. 6, 530–541 (2005).
Capila, I. & Linhardt, R.J. Heparin-protein interactions. Angew. Chem. Int. Edn Engl. 41, 391–412 (2002).
Sasisekharan, R., Shriver, Z., Venkataraman, G. & Narayanasami, U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat. Rev. Cancer 2, 521–528 (2002).
Bradbury, E.J. et al. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416, 636–640 (2002).
Kitagawa, H., Tsutsumi, K., Tone, Y. & Sugahara, K. Developmental regulation of the sulfation profile of chondroitin sulfate chains in the chicken embryo brain. J. Biol. Chem. 272, 31377–31381 (1997).
Plaas, A.H.K., West, L.A., Wong-Palms, S. & Nelson, F.R.T. Glycosaminoglycan sulfation in human osteoarthritis. Disease-related alterations at the non-reducing termini of chondroitin and dermatan sulfate. J. Biol. Chem. 273, 12642–12649 (1998).
Holt, C.E. & Dickson, B.J. Sugar codes for axons? Neuron 46, 169–172 (2005).
Nandini, C.D. et al. Structural and functional characterization of oversulfated chondroitin sulfate/dermatan sulfate hybrid chains from the notochord of hagfish. Neuritogenic and binding activities for growth factors and neurotrophic factors. J. Biol. Chem. 279, 50799–50809 (2004).
Ueoka, C. et al. Neuronal cell adhesion, mediated by the heparin-binding neuroregulatory factor midkine, is specifically inhibited by chondroitin sulfate E. Structural and functional implications of the over-sulfated chondroitin sulfate. J. Biol. Chem. 275, 37407–37413 (2000).
Shuo, T. et al. Developmental changes in the biochemical and immunological characters of the carbohydrate moiety of neuroglycan C, a brain-specific chondroitin sulfate proteoglycan. Glycoconj. J. 20, 267–278 (2004).
Tsuchida, K. et al. Appican, the proteoglycan form of the amyloid precursor protein, contains chondroitin sulfate E in the repeating disaccharide region and 4-O-sulfated galactose in the linkage region. J. Biol. Chem. 276, 37155–37160 (2001).
Deepa, S.S., Yamada, S., Zako, M., Goldberger, O. & Sugahara, K. Chondroitin sulfate chains on syndecan-1 and syndecan-4 from normal murine mammary gland epithelial cells are structurally and functionally distinct and cooperate with heparan sulfate chains to bind growth factors. A novel function to control binding of midkine, pleiotrophin, and basic fibroblast growth factor. J. Biol. Chem. 279, 37368–37376 (2004).
Karst, N.A. & Linhardt, R.J. Recent chemical and enzymatic approaches to the synthesis of glycosaminoglycan oligosaccharides. Curr. Med. Chem. 10, 1993–2031 (2003).
Tully, S.E. et al. A chondroitin sulfate small molecule that stimulates neuronal growth. J. Am. Chem. Soc. 126, 7736–7737 (2004).
Jacquinet, J.-C., Rochepeau-Jobron, L. & Combal, J.-P. Multigram syntheses of the disaccharide repeating units of chondroitin 4- and 6-sulfates. Carbohydr. Res. 314, 283–288 (1998).
Brickman, Y.G. et al. Structural modification of fibroblast growth factor-binding heparan sulfate at a determinative stage of neural development. J. Biol. Chem. 273, 4350–4359 (1998).
Kantor, D.B. et al. Semaphorin 5A is a bifunctional axon guidance cue regulated by heparan and chondroitin sulfate proteoglycans. Neuron 44, 961–975 (2004).
Mayo, S.L., Olafson, B.D. & Goddard, W.A., III DREIDING: A generic force field for molecular simulations. J. Phys. Chem. 94, 8897–8909 (1990).
Rappe, A.K. & Goddard, W.A., III Charge equilibration for molecular dynamics simulations. J. Phys. Chem. 95, 3358–3363 (1991).
Lim, K.-T. et al. Molecular dynamics for very large systems on massively parallel computers: the MPSim program. J. Comput. Chem. 18, 501–521 (1997).
Feizi, T., Fazio, F., Chai, W. & Wong, C.H. Carbohydrate microarrays—a new set of technologies at the frontiers of glycomics. Curr. Opin. Struct. Biol. 13, 637–645 (2003).
de Paz, J.L., Noti, C. & Seeberger, P.H. Microarrays of synthetic heparin oligosaccharides. J. Am. Chem. Soc. 128, 2766–2767 (2006).
Park, S., Lee, M.R., Pyo, S.J. & Shin, I. Carbohydrate chips for studying high-throughput carbohydrate-protein interactions. J. Am. Chem. Soc. 126, 4812–4819 (2004).
Muramatsu, T. Midkine and pleiotrophin: two related proteins involved in development, survival, inflammation and tumorigenesis. J. Biochem. 132, 359–371 (2002).
Herndon, M.E., Stipp, C.S. & Lander, A.D. Interactions of neural glycosaminoglycans and proteoglycans with protein ligands: assessment of selectivity, heterogeneity and the participation of core proteins in binding. Glycobiology 9, 143–155 (1999).
Huang, E.J. & Reichardt, L.F. Neurotrophins: roles in neuronal development and function. Annu. Rev. Neurosci. 24, 677–736 (2001).
Bicknese, A.R., Sheppard, A.M., O'Leary, D.D. & Pearlman, A.L. Thalamocortical axons extend along a chondroitin sulfate proteoglycan-enriched pathway coincident with the neocortical subplate and distinct from the efferent path. J. Neurosci. 14, 3500–3510 (1994).
Jiang, B., Akaneya, Y., Hata, Y. & Tsumoto, T. Long-term depression is not induced by low-frequency stimulation in rat visual cortex in vivo: a possible preventing role of endogenous brain-derived neurotrophic factor. J. Neurosci. 23, 3761–3770 (2003).
Seil, F.J. & Drake-Baumann, R. TrkB receptor ligands promote activity-dependent inhibitory synaptogenesis. J. Neurosci. 20, 5367–5373 (2000).
Skoff, A.M. & Adler, J.E. Nerve growth factor regulates substance P in adult sensory neurons through both TrkA and p75 receptors. Exp. Neurol. 197, 430–436 (2006).
Bellaiche, Y., The, I. & Perrimon, N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature 394, 85–88 (1998).
Fernaud-Espinosa, I., Nieto-Sampedro, M. & Bovolenta, P. Developmental distribution of glycosaminoglycans in embryonic rat brain: relationship to axonal tract formation. J. Neurobiol. 30, 410–424 (1996).
Greeley, B.H. et al. New pseudospectral algorithms for electronic structure calculations: length scale separation and analytical two-electron integral corrections. J. Chem. Phys. 101, 4028–4041 (1994).
Acknowledgements
We thank V.W.T. Kam for assistance with modifying the Dreiding force field; R.H. Grubbs for the catalyst; C.J. Rogers for assistance with data analysis; J.L. Riechmann, director of the Millard and Muriel Jacobs Genetic and Genomics Laboratory at Caltech, for assistance with printing the microarrays; S. Ou in the Caltech Monoclonal Antibody Facility; and the Biological Imaging Center and Environmental Analysis Center at Caltech for instrumentation. This work was supported by the American Cancer Society (RSG-05-106-01-CDD), Human Frontiers Science Program, Tobacco-Related Disease Research Program (14RT-0034), National Institutes of Health (RO1 NS045061, C.I.G. and L.C.H.-W.), National Science Foundation (S.E.T.) and Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
C.I.G. and S.E.T. contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Each CS tetrasaccharide favors a distinct set of torsion angles: torsion plots for ψ1 and φ1. (PDF 656 kb)
Supplementary Fig. 2
Each CS tetrasaccharide favors a distinct set of torsion angles: torsion plots for ψ2 and φ2. (PDF 586 kb)
Supplementary Fig. 3
Each CS tetrasaccharide favors a distinct set of torsion angles: torsion plots for ψ3 and φ3. (PDF 587 kb)
Supplementary Fig. 4
Each CS tetrasaccharide presents a unique van der Waals surface. (PDF 35 kb)
Supplementary Fig. 5
Monoclonal antibody 5D2-2C2 (raised against CS-C) selectively recognizes the CS-C tetrasaccharide. (PDF 44 kb)
Supplementary Fig. 6
Antibodies against BDNF or midkine, but not control IgG antibodies, block the binding of BDNF or midkine to CS-E on the microarray. (PDF 49 kb)
Supplementary Fig. 7
Function-blocking antibodies against BDNF and TrkB, but not class-matched control antibodies, disrupt the neuritogenic activity of CS-E. (PDF 47 kb)
Supplementary Methods
Experimental procedures for the synthesis of CS tetrasaccharides 1, 2, 3 and 4, conjugation methods for preparing carbohydrate microarrays, and dot blot procedures. (PDF 380 kb)
Rights and permissions
About this article
Cite this article
Gama, C., Tully, S., Sotogaku, N. et al. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol 2, 467–473 (2006). https://doi.org/10.1038/nchembio810
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio810
This article is cited by
-
Sulfated glycosaminoglycans inhibit transglutaminase 2 by stabilizing its closed conformation
Scientific Reports (2022)
-
Chemical editing of proteoglycan architecture
Nature Chemical Biology (2022)
-
Development of a bispecific immune engager using a recombinant malaria protein
Cell Death & Disease (2021)
-
The C-terminal peptide of CCL21 drastically augments CCL21 activity through the dendritic cell lymph node homing receptor CCR7 by interaction with the receptor N-terminus
Cellular and Molecular Life Sciences (2021)
-
Structural insight into the binding of human galectins to corneal keratan sulfate, its desulfated form and related saccharides
Scientific Reports (2020)