Abstract
The rapid identification of highly specific and potent drug candidates continues to be a substantial challenge with traditional pharmaceutical approaches. Moreover, many targets have proven to be intractable to traditional small-molecule and protein approaches. Therapeutics based on RNA interference (RNAi) offer a powerful method for rapidly identifying specific and potent inhibitors of disease targets from all molecular classes. Numerous proof-of-concept studies in animal models of human disease demonstrate the broad potential application of RNAi therapeutics. The major challenge for successful drug development is identifying delivery strategies that can be translated to the clinic. With advances in this area and the commencement of multiple clinical trials with RNAi therapeutic candidates, a transformation in modern medicine may soon be realized.
Similar content being viewed by others
Main
RNAi is a fundamental cellular mechanism for silencing gene expression that can be harnessed for the development of new drugs1,2. The reduction in expression of pathological proteins through RNAi is applicable to all classes of molecular targets, including those that are difficult to modulate selectively with traditional pharmaceutical approaches involving small molecules or proteins. Consequently, RNAi therapeutics as a drug class have the potential to exert a transformational effect on modern medicine. In RNAi, the target mRNA is enzymatically cleaved, leading to decreased abundance of the corresponding protein, and specificity is a key feature of the mechanism. Synthetic small interfering RNAs (siRNAs) leverage the naturally occurring RNAi process in a manner that is consistent and predictable with regard to extent and duration of action. In addition, viral delivery of short hairpin RNAs (shRNAs) represents an alternative strategy for harnessing RNAi. Both nonviral delivery of siRNAs and viral delivery of shRNAs are being advanced as potential RNAi-based therapeutic approaches.
In this review, we provide an overview of the molecular mechanism of RNAi; the in silico design of siRNAs and shRNAs that are specific for a target of interest, in the context of current concepts relating chemical structure to specificity and potency; the use of chemical modifications that confer stability against exo- and endonucleases present in biological fluids and tissues; and strategies for facilitating cellular delivery in vivo through conjugation, complexation and lipid-based approaches to facilitate cellular uptake. We summarize the numerous publications to date demonstrating the robust efficacy of RNAi in animal models of human disease upon direct (local) as well as systemic administration. These proof-of-concept studies support RNAi as the basis for a new therapeutic approach that has the potential to change the treatment of human disease. Most importantly, as we will discuss, clinical trials have recently commenced, with RNAi therapeutic candidates under study for treatment of age-related macular degeneration (AMD) and respiratory syncytial virus (RSV) infection.
Molecular mechanism of RNAi
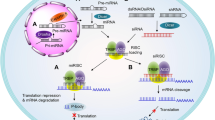
The RNase Dicer initiates RNAi by cleaving double-stranded RNA substrates into small fragments of about 21–25 nucleotides in length (Fig. 1). These siRNA duplexes are incorporated into a protein complex called the RNA-induced silencing complex (RISC; Fig. 1). Biochemical analysis identified Argonaute 2 (Ago2) as the protein in RISC responsible for mRNA cleavage3, and the crystal structure of RNA-bound Ago2 has been reported, revealing key interactions4.
Long double-stranded RNA (dsRNA) is cleaved, by the enzyme Dicer, into small interfering RNA (siRNA). These siRNAs are incorporated into the RNA-induced silencing complex (RISC), where the strands are separated. The RISC containing the guide or antisense strand seeks out and binds to complementary mRNA sequences. These mRNA sequences are then cleaved by Argonaute, the enzyme within the RISC responsible for mRNA degradation, which leads to mRNA down-modulation. A, adenosine.
Before RISC activation, the sense (nonguide) strand of the siRNA duplex is cleaved by Ago2, in the same manner as it cleaves mRNA substrates5,6. Preventing sense-strand cleavage by chemical modification can reduce siRNA potency in vitro; however, experimental context is important, as siRNAs with highly stabilized (uncleavable) sense strands can be highly active.
Role of chemical modifications
Small-molecule pharmaceutical drugs, almost without exception, meet the 'Lipinski Rules', criteria including high lipophilicity and molecular weight of not more than 500. In sharp contrast, siRNAs naturally lack these drug-like properties owing to their large size (two turns of a nucleic acid double helix), nearly 40 anionic charges due to the phosphodiester backbone, and high molecular weight (over 13 kDa). In aqueous solution, with their sugar-phosphate backbone exposed to water, siRNAs are extremely hydrophilic and heavily hydrated. Furthermore, siRNAs are unstable in serum as a result of degradation by serum nucleases, contributing to their short half-lives in vivo7. Although the molecular weight of siRNAs cannot be reduced, these molecules can be made more 'drug-like' through judicious use of chemical modification to the sugars, backbone or bases of the oligoribonucleotides.
Chemically modified siRNA duplexes have been evaluated in cell-based assays and in animal models. The modifications discussed are shown in Figure 2. Stability against nuclease degradation has been achieved by introducing a phosphorothioate (P=S) backbone linkage at the 3′ end for exonuclease resistance and 2′ modifications (2′-OMe, 2′-F and related) for endonuclease resistance8,9,10. An siRNA motif, consisting entirely of 2′-O-methyl and 2′-fluoro nucleotides, has enhanced plasma stability and increased in vitro potency. At one site, this motif shows >500-fold improvement in potency over the unmodified siRNA11. Using phosphatase and tensin homolog (PTEN) as a target, the effect of 2′ sugar modifications such as 2′-fluoro-2′-deoxynucleoside (2′-F), 2′-O-methyl (2′-O-Me) and 2′-O-(2-methoxyethyl) (2′-O-MOE) in the guide and nonguide strands was evaluated in HeLa cells. The activity depends on the position of the modification in the guide-strand sequence. The siRNAs with modified residues at the 5′ end of the guide strand seem to be less active than those modified at the 3′ end. The 2′-F sugar is generally well tolerated on the guide strand, whereas the 2′-O-MOE modification results in loss of activity regardless of placement position in the construct. The incorporation of 2′-O-Me and 2′-O-MOE in the nonguide strand of siRNA does not have a notable effect on activity12. Sugar modifications such as 2′-O-Me, 2′-F and locked nucleic acid (LNA, with a methylene bridge connecting 2′ and 4′ carbons) seem to be able to reduce the immunostimulatory effects of siRNAs (see below).
Duplexes containing the 4′-thioribose modification have a stability 600 times greater than that of natural RNA13. Crystal structure studies reveal that 4′-thioriboses adopt conformations very similar to the C3′-endo pucker observed for unmodified sugars in the native duplex14. Stretches of 4′-thio-RNA were well tolerated in both the guide and nonguide strands. However, optimization of both the number and the placement of 4′-thioribonucleosides is necessary for maximal potency. These optimized siRNAs are generally equipotent with or superior to native siRNAs and show increased thermal and plasma stability. Furthermore, substantial improvements in siRNA activity and plasma stability have been achieved by judicious combination of 4′-thioribose with 2′-O-Me and 2′-O-MOE modifications15.
As mentioned, phosphorothioate (P=S) modifications are generally well tolerated on both strands and provide improved nuclease resistance. The 2′,5′-phosphodiester linkages seem to be tolerated in the nonguide but not the guide strand of the siRNA16. In the boranophosphate linkage, a nonbridging phosphodiester oxygen is replaced by an isoelectronic borane (BH3-) moiety. Boranophosphate siRNAs have been synthesized by enzymatic routes using T7 RNA polymerase and a boranophosphate ribonucleoside triphosphate in the transcription reaction. Boranophosphate siRNAs are more active than native siRNAs if the center of the guide strand is not modified, and they may be at least ten times more nuclease resistant than unmodified siRNAs17,18.
siRNA duplexes containing the 2,4-difluorotoluyl ribonucleoside (rF) were synthesized to evaluate the effect of noncanonical nucleoside mimetics on RNA interference. Thermal melting analysis showed that the base pair between rF and adenosine is destabilizing relative to a uridine-adenosine pair, although it is slightly less destabilizing than other mismatches. The crystal structure of a duplex containing rF-adenosine pairs shows local structural variations relative to a canonical RNA helix. As the fluorine atoms cannot act as hydrogen bond acceptors and are more hydrophobic than uridine, a well-ordered water structure is not seen around the rF residues in both grooves. Rapid amplification of 5 complementary DNA ends (5′-RACE) analysis confirms cleavage of target mRNA opposite to the rF placement site19,20.
Certain terminal conjugates have been reported to improve or direct cellular uptake. For example, siRNAs conjugated with cholesterol improve in vitro and in vivo cell permeation in liver cells6. As described below, cholesterol and an RNA aptamer conjugation show promise in animal models.
Design considerations for potency and specificity
Critical design concerns in the selection of siRNA duplexes for therapeutic use are potency and specificity. There are two major considerations with regard to siRNA specificity: 'off-targeting' due to silencing of genes sharing partial homology with the siRNA, and 'immune stimulation' due to the engagement of components of the innate immune system by the siRNA duplex. A combination of bioinformatics methods, chemical modification strategies and empirical testing is required to address these issues.
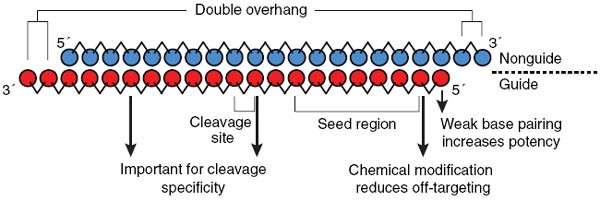
Concomitant with the first description of the structure of active siRNAs, a set of 'rules' was proposed for selecting potent siRNA duplex sequences21,22. Several groups have subsequently developed more sophisticated extensions of these largely empirical criteria, leading to the development of algorithms for siRNA design23,24. Recent biochemical studies of the molecular mechanism of RNA interference have highlighted some key features of potent siRNA duplexes (Fig. 3). Most notably, it has been found that the efficiency with which the guide strand is incorporated into the RISC complex is perhaps the most important factor determining siRNA potency. Because siRNA duplexes are symmetric, the question arose of how the RISC machinery is able to determine which strand to use for target silencing. Insight into this enigma came from careful analyses of microRNAs (miRNAs), the endogenous counterparts of siRNAs. Examination of the sequences of a large number of vertebrate and invertebrate miRNA precursor sequences showed that the predicted thermodynamic stabilities of the two ends of the duplex are unequal25,26. Specifically, calculating the ΔG for the several base pairs at each end of the duplex revealed that the 5′ end of the mature miRNA pairs less tightly with the carrier strand than does the 3′ end. In short, miRNA precursors show thermodynamic asymmetry. It was hypothesized that components of the RISC machinery select the guide strand based on this asymmetry.
Experimental evidence supporting the asymmetry hypothesis has been derived from studies using chemically synthesized siRNAs in transfection experiments. Through an elegant assay in which each strand of the siRNA targets a different reporter gene, Schwarz et al. were able to quantify the relative efficiency of RISC incorporation for each of the two strands25. They found that the RISC machinery preferentially incorporates the strand whose 5′ end binds less tightly with the other strand. In fact, strand selection could be switched by making a single nucleotide substitution at the end of the duplex to alter relative binding of the ends. A similar conclusion was reached by another group based on in vitro screening of a large collection of siRNAs with varying potency26,27. Thus, designing siRNAs with relatively weaker base pairing at the 5′ end of the desired guide strand may increase the likelihood of obtaining a potent duplex.
The issue of off-target silencing has been the subject of intensive study in a number of different laboratories over the past several years. Transcriptional profiling studies have confirmed that siRNA duplexes can potentially silence multiple genes in addition to the intended target. As expected, genes in these so called off-target 'signatures' contain regions that are complementary to one of the two strands in the siRNA duplex28,29,30. More detailed bioinformatic analyses have revealed that the regions of complementarity are most often found in the 3′ UTRs of the off-target genes31. This immediately suggested a microRNA-like mechanism, because miRNAs generally interact with the 3′ UTR region of their targets. Evidence in support of this concept came from a closer look at the determinants of siRNA off-targeting. It was discovered that sequence complementarity between the 5′ end of the guide strand and the mRNA is the key to off-target silencing31,32. The critical nucleotides were found to be positions 2–8, counting from the 5′ end of the guide strand (Fig. 3). This corresponds to the so-called 'seed region' of miRNAs, which has been shown to determine miRNA specificity33.
Two strategies for avoiding seed region–mediated off-targeting can be envisioned. The first is simply to ensure that nucleotides complementary to positions 2–8 of the guide strand are unique to the intended target. Though theoretically possible, this approach may prove impractical, as the universe of possible seed-region heptamers is only 16,384 distinct sequences. Even if the homology is restricted to the 3′ UTR, it may prove difficult to identify siRNA duplexes satisfying the criteria of potency and specificity. As one alternative, recent published work has reported that off-targeting can be substantially reduced by chemical modification of nucleotides within the seed region34. Specifically, the introduction of a 2′-O-Me modification into nucleotides within the seed region was shown to inactivate the off-target activity of the siRNA without compromising silencing of the intended mRNA. In fact, introduction of the modification at a single nucleotide position (position 2, Fig. 3) is sufficient to suppress the majority of off-targeting. The mechanism, anticipated by recently published crystal structure data, appears to involve perturbation of RISC interaction with the modified nucleotide.
Interactions outside of the seed region can also substantially affect siRNA specificity. Although the seed region seems to be critical for mRNA recognition, notable mRNA cleavage requires more extensive base pairing between the siRNA and the target32. In a recent study, Schwarz et al. designed siRNAs capable of distinguishing between mRNA targets that differ by only one nucleotide35. They showed that target selectivity depends on the location of the mismatch between the siRNA and the mRNA. Whereas positioning the mismatch within the seed region imparts a certain degree of selectivity, positioning the mismatch further 3′ in the guide strand (especially at positions 10 and 16, Fig. 3) produces highly discriminatory siRNAs. The authors hypothesized that mismatches at these positions are particularly disruptive to the helical structure of the siRNA–mRNA complex required for target cleavage.
A second mechanism whereby siRNA duplexes can induce unintended effects is through stimulation of the innate immune system in certain specialized immune cell types. It has been demonstrated that siRNA duplexes harboring distinct sequence motifs can engage Toll-like receptors (TLRs) in plasmacytoid dendritic cells, resulting in increased production of interferon36. Such immune stimulation could pose a significant problem in a therapeutic setting. This phenomenon is reminiscent of the results of earlier studies with DNA antisense oligonucleotides in which distinct sequences (so-called CpG motifs) were shown to be immunostimulatory37. Subsequent studies established that TLR-9, the receptor for unmethylated CpG-containing pathogen DNA, is the innate immune regulator engaged by antisense oligonucleotides38. In the case of siRNAs, it seems to be TLR-7 that is the mediator of immune stimulation36.
Several possible strategies exist for avoiding immune stimulation by siRNA duplexes, including avoidance of the offending sequences during siRNA design and chemical modification to inactivate the motifs. The former approach is not feasible at present because the full spectrum of stimulatory motifs has not been identified. Evidence supporting the latter approach comes from studies in which chemical modifications at the 2′ position of nucleotides within putative TLR-7–interacting sequences eliminate immune stimulation without compromising silencing activity36,39. Another possibility would be to use siRNA delivery strategies that avoid the cell types responsible for immune stimulation.
Prediction of the nucleotide sequence and chemical modifications required to yield an ideal siRNA duplex remains a work in progress. Still, the recent advances described above have allowed the development of design algorithms that greatly increase the likelihood of success. It is nonetheless important to note that the relevance of in vitro measurements of potency and specificity to in vivo activity in a therapeutic setting has yet to be established. For example, the spectrum of off-target genes identified in tissue culture studies can differ depending on the method by which siRNAs are introduced into cells40. Also, the induction of an innate immune response by certain siRNA sequences is cell type dependent41. At present, the most prudent and robust strategy is to synthesize and screen a substantial library of siRNA duplexes for each target of interest (perhaps even 'tiling' the entire messenger RNA) to identify the most promising candidates.
Proof of concept for local RNAi in animal models
During the past several years, numerous studies have been published demonstrating efficacious silencing of disease genes by local administration of siRNAs or shRNAs in animal models of human disease. Both exogenous and endogenous genes have been silenced, and promising in vivo results have been obtained across multiple organs and tissues. Efficacy has been demonstrated for viral infection (respiratory and vaginal), ocular disease, disorders of the nervous system, cancer and inflammatory bowel disease (Fig. 4). An important aspect of these proof-of-concept studies is that they have supported the expected high specificity of RNAi.
Direct RNAi represents local delivery of RNAi, and has been carried out successfully to specific tissues or organs, including lung, eye, the nervous system, tumors, the digestive system and vagina. Systemic RNAi represents intravenous delivery of RNAi and has been carried out successfully to lung, tumors, liver and joint. Specific disease models are indicated where efficacy was achieved.
Local RNAi can protect against both respiratory42,9and vaginal43 viral infections. Two reports illustrate efficacious direct delivery of siRNA to the lung in rodent and monkey models of RSV, influenza and severe acute respiratory syndrome (SARS) infection with and without lipid formulation. In mouse models of infection, pulmonary viral titers of RSV and parainfluenza were reduced by more than 99% with intranasal delivery of siRNAs formulated with TransIT-TKO, a cationic polymer–based transfection reagent, targeting RSV and parainfluenza virus, respectively42. In addition, siRNA targeting RSV reduced pulmonary pathology, as assessed by respiratory rate, leukotriene induction and inflammation. These positive proof-of-concept studies in mice have led to clinical trials of RNAi therapeutics targeting RSV.
Another system for which there have been multiple examples of efficacious local delivery of siRNA is the eye, where proof of concept has been successfully achieved in animal models of ocular neovascularization and scarring using saline and lipid formulations44,45,46. Intravitreal injection of siRNA targeting vascular endothelial growth factor (VEGF) receptor-1, formulated in phosphate-buffered saline, was effective in reducing the area of ocular neovascularization by one-third to two-thirds in two mouse models44. In addition, siRNAs targeting VEGF and the transforming growth factor-β receptor type II, formulated with TransIT-TKO, were injected directly into the mouse eye, resulting in inhibition of laser photocoagulation–induced choroidal neovascularization45 and latex bead–induced collagen deposition and inflammatory cell infiltration46, respectively. As with the lung, multiple siRNA formulations were effective in the eye. These encouraging proof-of-concept studies in animal models have led to clinical trials of siRNAs targeting the VEGF pathway in AMD.
In the nervous system, RNAi has been particularly useful for validating disease targets in vivo. Again, several formulations, including saline, polymer complexation and lipid or liposomal formulations, have been efficacious for delivering siRNAs locally to the nervous system in numerous disease models. The simplest mode of delivery is intracerebroventricular, intrathecal or intraparenchymal infusion of naked siRNA formulated in buffered isotonic saline, which results in silencing of specific neuronal molecular mRNA targets in multiple regions of the central and peripheral nervous systems47,48,49,50. With naked siRNA formulated in buffered isotonic saline, doses of 0.4 mg per day are typically required for effective target gene silencing. Polymer complexation and lipid or liposomal formulations such as polyethylene imine (PEI), iFECT, DOTAP and JetSI/DOPE facilitate cellular uptake and reduce the doses of siRNA required for effective neuronal target silencing in vivo to approximately 5–40 μg51,52,53,54.
Local viral delivery of shRNA to the nervous system has been reported in vivo with adenoviral, adeno-associated viral (AAV) and lentiviral delivery in normal mice55 as well as in animal models of spinocerebellar ataxia56, Huntington disease57,58, amyotrophic lateral sclerosis (ALS)59,60 and Alzheimer disease61, where abnormal, disease phenotypes including behavior and neuropathology were normalized. Notably, all of the in vivo studies to date have targeted genes expressed in neurons; it remains to be seen whether silencing in vivo can be achieved in other nervous-system cell types such as oligodendrocytes and astrocytes. Moreover, for endogenous neuronal targets, expression of the target gene is typically reduced only partially, and in some cases by as little as 10–20%, yet this modest reduction in mRNA results in a marked effect on the specific behavior appropriate to the targeted gene.
For application to oncology, direct delivery of siRNAs and viral delivery of shRNAs to tumors have been successful in inhibiting xenograft growth in several mouse models. A number of approaches—including lipid-based formulation (TransMessenger62) and complexation with PEI63, cholesterol-oligoarginine64, a protamine-Fab fusion protein65 and atelocollagen66,67—have been shown to facilitate delivery into tumor cells. Notably, these siRNA delivery approaches are effective with several or even a single intratumor injection of siRNA, at microgram doses. Very recently, aptamer-siRNA chimeric RNAs have also been used successfully to facilitate siRNA delivery in vivo, resulting in tumor regression in a xenograft model of prostate cancer68. Viral and vector-based delivery of shRNAs directly to the tumor site69,70 has also been used effectively in mouse models of adenocarcinoma, Ewing sarcoma and prostate cancer. Of the multiple delivery strategies that have been effective in mouse tumor models, the aptamer approach has the potential of substantially simplifying delivery, if an aptamer is available for a tumor-specific receptor such as prostate-specific membrane antigen (PMSA) and the large-scale synthesis of such a construct is feasible.
For inflammatory bowel disease, direct delivery of siRNA targeting tumor necrosis factor-α (TNF-α) with a Lipofectamine formulation has recently been shown to reduce not only TNF-α abundance but also colonic inflammation after administration by enema71. This report, together with a study of siRNA targeting herpes simplex virus-2 (ref. 43), suggests that mucosal surfaces are accessible with liposomal siRNA formulations.
Proof of concept for systemic RNAi in animal models
Over the past several years, a number of studies have been published demonstrating the silencing of disease genes by systemic administration of siRNAs (Fig. 4; reviewed in refs. 1,72). In some of these studies, silencing of endogenously expressed genes has shown promising in vivo results in different disease contexts. For example, efficacy has been demonstrated in mouse models of hypercholesterolemia and rheumatoid arthritis. In other work, systemic RNAi targeting exogenous genes has shown promise in models of viral infection (hepatitis B virus (HBV), influenza virus, Ebola virus) and in tumor xenografts. Critical to the success of most of these studies has been the use of chemical modifications or delivery formulations that impart desirable pharmacokinetic properties to the siRNA duplex and that also promote cellular uptake in tissues.
In 2004, Soutschek et al. demonstrated effective silencing of the apolipoprotein apoB in mice by intravenous administration of cholesterol-conjugated siRNA duplexes6. Three daily injections of cholesterol-conjugated siRNA at a dose of 50 mg kg−1 resulted in silencing of the apoB mRNA by 57% and 73%, respectively, in the liver and jejunum, the two principal sites for apoB expression. The mechanism of action was proven, by 5′-RACE, to be RNAi mediated. This mRNA silencing produced a 68% reduction in apoB protein abundance in plasma and a 37% reduction in total cholesterol. These therapeutically relevant findings were completely consistent with the known function of apoB in lipid metabolism. Cholesterol conjugation imparted critical pharmacokinetic and cellular uptake properties to the siRNA duplex.
Further advances in systemic RNAi with optimized delivery have recently been reported. Recently, Zimmermann et al. made use of siRNA duplexes formulated in stable nucleic acid lipid particles (SNALPs39) to recapitulate the silencing of apoB in mice73. In rodent studies, the silencing produced by a single dose of SNALP-formulated siRNA at 2.5 mg kg−1 was greater than that reported in the earlier study using cholesterol-conjugated siRNAs. More importantly, therapeutic silencing of apoB was also demonstrated in nonhuman primates. A single dose of 2.5 mg kg−1 siRNA encapsulated in the SNALP formulation reduced apoB mRNA in the livers of cynomolgus monkeys by more than 90%. As in the mouse experiments, apoB silencing was accompanied by substantial reductions in serum cholesterol (>65%) and low-density lipoproteins (>85%). Furthermore, silencing was shown to last for at least 11 d after a single dose. In addition, the treatment seemed to be well tolerated, with transient increases in liver enzymes as the only reported evidence of toxicity. This primate study represented an important step forward in the development of systemic RNAi for therapeutic applications. Moreover, the general applicability of SNALP formulations for hepatic delivery of siRNA has been demonstrated in animal models of HBV and Ebola virus infection74,75.
In mouse tumor xenograft models, the efficacy of systemic RNAi has been demonstrated using a variety of delivery strategies (reviewed in refs. 1,2,76). Systemically delivered cationic cardiolipin liposomes containing siRNA specific for Raf-1 inhibit tumor growth in a xenograft model of human prostate cancer77. Vascular endothelial growth factor receptor-2 (VEGF-R2)-targeting siRNAs complexed with self-assembling nanoparticles consisting of polyethylene glycol-conjugated (PEGylated) PEI with an Arg-Gly-Asp peptide attached at the distal end of the PEG accumulate in tumors and cause inhibition of VEGF-R2 expression. Intravenous administration of these complexes into tumor-bearing mice inhibits both tumor angiogenesis and growth rate78. Simpler PEI formulations have also shown efficacy in xenograft tumor models79, as have complexes of siRNA duplexes with atelocollagen. Systemic administration of atelocollagen–siRNA complexes has marked effects on subcutaneous tumor xenografts66 as well as bone metastases76. Another recently described delivery strategy made use of a recombinant antibody fusion protein to achieve cell type–specific delivery. As described above, Song et al. fused the nucleic acid binding protein protamine to the C terminus of a fragment antibody (Fab) targeting the HIV-1 envelope protein gp160. After systemic administration, the Fab-protamine fusion was able to deliver an siRNA mixture to mouse melanoma cells engineered to express the envelope protein, leading to substantial inhibition of tumor growth in mice. Tumors derived from cells not expressing the envelope protein were unaffected. In another example of ligand-directed delivery, Hu-Lieskovan et al. made use of transferrin-conjugated nanoparticles to deliver an siRNA targeting the oncogenic EWS-FLI translocation-derived mRNA in a mouse model of metastatic Ewing sarcoma80. Removal of the targeting ligand or the use of a control siRNA sequence eliminated the antitumor effects.
Comparison of different delivery strategies for RNAi
Effective delivery is perhaps the most challenging remaining consideration for successful translation of RNAi to the clinic and to broad use in patients. In the animal studies reviewed above, nonviral and viral approaches, local and systemic administration, and multiple formulations (saline, lipids, and complexes or conjugates with small molecules, polymers, proteins and antibodies) have all been used to achieve efficacy. However, each of these approaches has distinct advantages and disadvantages for clinical translation, which require careful consideration.
Although viral delivery provides the potential advantage that a single administration could lead to durable down-modulation of the targeted pathological protein, a major risk was highlighted recently81. With AAV delivery of shRNAs, excessive diversion of the endogenous RNAi mechanism occurred that resulted in pronounced toxicity in mice. Clearly, for all drugs, it is critical to be able to control the level of drug and the duration of drug action, such that the exposure is safe while still being efficacious. In distinct contrast to nonviral delivery of siRNAs, a substantial liability of viral delivery is that it is impossible to fully predict drug exposure, with regard to both amount and timing. In addition, as shRNAs enter the RNAi pathway upstream of therapeutic applications for siRNAs, viral vectors expressing high levels of shRNAs may interfere with endogenous miRNA biogenesis.
The principal considerations for selecting local versus systemic siRNA administration are the doses needed to achieve sufficient drug concentration in the target tissue and the possible effects of the exposure of nontargeted tissues to drug. At one extreme, with certain tissues, efficacy has so far been demonstrated only with local delivery; current formulations may not provide sufficient drug concentration in the target tissue after systemic delivery. However, with other tissues (for example, liver), intravenous doses in the low mg kg−1 range with liposomal formulation provide robust therapeutic gene silencing. In general, and as with any pharmacologic approach, the doses of siRNA required for efficacy are substantially lower when siRNAs are injected into or near the target tissue than when they are administered systemically. Given the high specificity of siRNAs for their intended molecular target, exposure of nontargeted tissues to drug is an issue only if the molecular target is expressed in nontargeted tissue and has an important role in normal cellular function within that tissue. In these cases, local delivery with more focused exposure might circumvent undesired side effects resulting from systemic delivery.
Liposomes, and lipid complexes or conjugates with small molecules, polymers, proteins and antibodies, have all been used to facilitate delivery of siRNAs to target cells. With these delivery partners, more robust efficacy can be achieved with doses of siRNA that are substantially lower, less frequent or both. For the additional (non-siRNA) components, however, there are associated biological and large-scale manufacturing considerations. Lipids and polymers can have cytotoxic effects that might limit their use in siRNA delivery for particular disease indications and dosing paradigms. However, it seems to be possible to identify lipid-based formulations and dosing regimens for which cytotoxicity is minimal and the risk of histopathology is reduced51,52,53. Small molecules, proteins and antibodies used as conjugates also need to be considered from the standpoint of biological activity. If the endogenous molecule (for example, receptor) with which they interact has an important role in normal physiology, then using this endogenous molecule to potentiate delivery may alter its normal function and produce undesired side effects. In all of these cases, the additional non-siRNA molecule or molecules increase the complexity of manufacturing, particularly at large scale. With scientific and technical advances, these approaches may provide marked enhancements to siRNA delivery with acceptable biological and manufacturing considerations.
Clinical trials with RNAi therapeutics
The first clinical trials with RNAi therapeutics target the VEGF pathway for the wet form of AMD and the RSV genome for treatment of RSV infection; in both cases the initial approach is direct administration of the RNAi therapeutic in a saline formulation. In both these cases, highly validated disease targets are being inhibited with siRNAs. Furthermore, direct administration of siRNAs to the eye and lung for AMD and RSV infection, respectively, maximizes the chances of delivering sufficient and therapeutically relevant concentrations of drug to the tissue of interest. In a phase 2 trial in patients with serious progressive AMD, the siRNA Cand5, targeting VEGF, has been reported to provide dose-related benefits with respect to several endpoints, including near vision and lesion size (http://www.acuitypharma.com/press/release13.pdf). Cand5 is also being tested for efficacy against diabetic macular edema in a phase 2 trial that began in early 2006 (http://www.acuitypharma.com/press/release10.pdf). The siRNA Sirna-027, targeting VEGF receptor-1, has recently completed phase 1 trials in patients with the wet form of AMD, in whom it was reported to be well tolerated. In addition, it was reported to stabilize or improve visual acuity in a subset of patients (http://www.sirna.com/wt/page/ocular). For RSV infection, two phase 1 trials with the siRNA ALN-RSV01 have been completed in over 100 healthy adult volunteers, in one of the largest human studies with an RNAi therapeutic, and ALN-RSV01 was found to be safe and well tolerated (http://phx.corporate-ir.net/phoenix.zhtml?c=148005&p=irol-newsArticle2&ID=849576&highlight=). Additional RNAi therapeutic candidates that are expected to advance into the clinic within the coming year include siRNAs targeting pandemic influenza (http://www.alnylam.com/therapeutic-programs/programs.asp) and hepatitis C (Sirna-034; http://www.sirna.com/wt/page/anti_viral). As these and other trials advance through the clinic in the near future, the exciting potential of siRNAs may be demonstrated.
Comparison of RNAi with traditional pharmaceutical drugs
As a therapeutic approach, RNAi provides solutions to the major drawbacks of traditional pharmaceutical drugs (Table 1). The principal advantages of RNAi over small-molecule and protein therapeutics are that all targets, including 'non-druggable' targets, can be inhibited with RNAi and that lead compounds can be rapidly identified and optimized. The primary challenge associated with small-molecule drugs is the identification of highly selective and potent compounds—a difficult and time-consuming process that, for some targets, can be unsuccessful. With RNAi, the identification of highly selective and potent sequences is rapid and has been demonstrated with numerous molecular targets across all molecular classes. With protein and antibody drugs, the main technical challenge is production. For proteins, acceptable cellular production levels are often difficult to achieve. For biologics as a therapeutic class, aggregation continues to be a major issue. In contrast, siRNAs are synthetic and easy to produce from a chemistry standpoint. Of course, with RNAi, by definition, only antagonism of the specific molecular target is possible, whereas small molecules, proteins and antibodies provide an opportunity for agonism of a molecular target. Overall, however, RNAi holds great promise as a therapeutic approach providing a major new class of drugs that will fill a significant gap in modern medicine.
Conclusions
Significant progress has been made in advancing RNAi therapeutics in a remarkably short period of time. Starting from the discovery that RNAi is mediated by long double-stranded RNA in Caenorhabditis elegans by Fire and Mello in 199882 and the publication in 2001 by Tuschl and his Max Planck Institute colleagues that synthetic siRNAs can silence target genes in mammalian systems20, the relatively short years since have seen an explosion in reports on therapeutic applications that harness RNAi. Clearly, the principal challenge that remains in achieving the broadest application of RNAi therapeutics is the hurdle of delivery. That said, tremendous progress has been made with new conjugation, complexation and lipid-based approaches, although the challenge of siRNA delivery has yet to be solved for all cell types in all organs. Once that challenge is met, the development of RNAi therapeutics will be limited primarily by target validation. It will then be possible to rapidly advance RNAi therapeutics against potentially any disease target in clinical studies and to thereby treat disease in a new manner. In the near future, the ongoing clinical trials with siRNAs for macular degeneration and RSV may reveal the exciting potential of RNAi therapeutics as the next major class of drug molecules.
References
Dykxhoorn, D.M., Palliser, D. & Lieberman, J. The silent treatment: siRNAs as small molecule drugs. Gene Ther. 13, 541–552 (2006).
Dykxhoorn, D.M. & Lieberman, J. Running interference: prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu. Rev. Biomed. Eng. 8, 377–402 (2006).
Rand, T.A., Ginalski, K., Grishin, N.V. & Wang, X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. USA 101, 14385–14389 (2004).
Ma, J.B. et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670 (2005).
Matranga, C., Tomari, Y., Shin, C., Bartel, D.P. & Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123, 607–620 (2005).
Rand, T.A., Petersen, S., Du, F. & Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123, 621–629 (2005).
Soutschek, J. et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432, 173–178 (2004).
Vornlocher, H.P., Zimmermann, T.S., Manoharan, M., Rajeev, KG., Roehl, I. & Akinc, A. Nuclease-resistant double-stranded RNA for RNA interference. Patent Cooperation Treaty International Application WO2005115481 (2005).
Li, B. et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 11, 944–951 (2005).
Choung, S., Kim, Y.J., Kim, S., Park, H.O. & Choi, Y.C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 342, 919–927 (2006).
Allerson, C.R. et al. Fully 2'-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 48, 901–904 (2005).
Prakash, T.P. et al. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 48, 4247–4253 (2005).
Hoshika, S., Minakawa, N. & Matsuda, A. Synthesis and physical and physiological properties of 4'-thioRNA: application to post-modification of RNA aptamer toward NF-κB. Nucleic Acids Res. 32, 3815–3825 (2004).
Haeberli, P., Berger, I., Pallan, P.S. & Egli, M. Syntheses of 4'-thioribonucleosides and thermodynamic stability and crystal structure of RNA oligomers with incorporated 4'-thiocytosine. Nucleic Acids Res. 33, 3965–3975 (2005).
Dande, P. et al. Improving RNA interference in mammalian cells by 4'-thio-modified small interfering RNA (siRNA): effect on siRNA activity and nuclease stability when used in combination with 2'-O-alkyl modifications. J. Med. Chem. 49, 1624–1634 (2006).
Prakash, T.P. et al. RNA interference by 2′,5′- linked nucleic acid duplexes in mammalian cells. Bioorg. Med. Chem. Lett. 16, 3238–3240 (2006).
Hall, A.H., Wan, J., Shaughnessy, E.E., Ramsay Shaw, B. & Alexander, K.A. RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucleic Acids Res. 32, 5991–6000 (2004).
Hall, A.H. et al. High potency silencing by single-stranded boranophosphate siRNA. Nucleic Acids Res. 34, 2773–2781 (2006).
Xia, J. et al. Gene silencing activity of siRNAs with a ribo-difluorotoluyl nucleotide. ACS Chem. Biol. 1, 176–183 (2006).
Somoza, A., Chelliserrykattil, J. & Kool, E.T. The roles of hydrogen bonding and sterics in RNA interference. Angew. Chem. Int. Edn. Engl. 45, 4994–4997 (2006).
Elbashir, S.M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Elbashir, S.M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001).
Boese, Q. et al. Mechanistic insights aid computational short interfering RNA design. Methods Enzymol. 392, 73–96 (2005).
Yuan, B., Latek, R., Hossbach, M., Tuschl, T. & Lewitter, F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 32, W130–W134 (2004).
Schwarz, D.S. et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003).
Khvorova, A., Reynolds, A. & Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003).
Reynolds, A. et al. Rational siRNA design for RNA interference. Nat. Biotechnol. 22, 326–330 (2004).
Jackson, A.L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 (2003).
Lin, X. et al. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 33, 4527–4535 (2005).
Qiu, S., Adema, C.M. & Lane, T. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 33, 1834–1847 (2005).
Jackson, A.L. et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187 (2006).
Birmingham, A. et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204 (2006).
Lewis, B.P., Burge, C.B. & Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Jackson, A.L. et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12, 1197–1205 (2006).
Schwarz, D.S. et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genetics (in the press).
Hornung, V. et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 11, 263–270 (2005).
Boggs, R.T. et al. Characterization and modulation of immune stimulation by modified oligonucleotides. Antisense Nucleic Acid Drug Dev. 7, 461–471 (1997).
Hemmi, H. et al. A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 (2000).
Judge, A.D. et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 23, 457–462 (2005).
Fedorov, Y. et al. Different delivery methods—different expression profiles. Nat. Methods 2, 241 (2005).
Reynolds, A. et al. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA 12, 988–993 (2006).
Bitko, V., Musiyenko, A., Shulyayeva, O. & Barik, S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 11, 50–55 (2005).
Palliser, D. et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature 439, 89–94 (2005).
Shen, J. et al. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 13, 225–234 (2006).
Reich, S.J. et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol. Vis. 9, 210–216 (2003).
Nakamura, H. et al. RNA interference targeting transforming growth factor-beta type II receptor suppresses ocular inflammation and fibrosis. Mol. Vis. 10, 703–711 (2004).
Makimura, H., Mizuno, T.M., Mastaitis, J.W., Agami, R. & Mobbs, C.V. Reducing hypothalamic AGRP by RNA interference increases metabolic rate and decreases body weight without influencing food intake. BMC Neurosci. 3, 18 (2002).
Thakker, D.R. et al. Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proc. Natl. Acad. Sci. USA 101, 17270–17275 (2004).
Thakker, D.R. et al. siRNA-mediated knockdown of the serotonin transporter in the adult mouse brain. Mol. Psychiatry 10, 782–789, 714 (2005).
Dorn, G. et al. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 32, e49 (2004).
Tan, P.H., Yang, L.C., Shih, H.C., Lan, K.C. & Cheng, J.T. Gene knockdown with intrathecal siRNA of NMDA receptor NR2B subunit reduces formalin-induced nociception in the rat. Gene Ther. 12, 59–66 (2005).
Luo, M.C. et al. An efficient intrathecal delivery of small interfering RNA to the spinal cord and peripheral neurons. Mol. Pain 1, 29 (2005).
Salahpour, A., Medvedev, I.O., Beaulieu, J.-M., Gainedinov, R.R. & Caron, M.G. Local knockdown of genes in the brain using small interfering RNA: a phenotypic comparison with knockout animals. Biol. Psychiatry, published online 17 May 2006.
Kumar, P., Lee, S.K., Shankar, P. & Manjunath, N. A single siRNA suppresses fatal encephalitis induced by two different flaviviruses. PLoS Med. 3, e96 (2006).
Chen, Y. et al. Adenovirus-mediated small-interference RNA for in vivo silencing of angiotensin AT1a receptors in mouse brain. Hypertension 47, 230–237 (2006).
Xia, H. et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 10, 816–820 (2004).
Harper, S.Q. et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl. Acad. Sci. USA 102, 5820–5825 (2005).
Rodriguez-Lebron, E., Denovan-Wright, E.M., Nash, K., Lewin, A.S. & Mandel, R.J. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol. Ther. 12, 618–633 (2005).
Ralph, G.S. et al. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat. Med. 11, 429–433 (2005).
Raoul, C. et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat. Med. 11, 423–428 (2005).
Singer, O. et al. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 8, 1343–1349 (2005).
Niu, X.Y., Peng, Z.L., Duan, W.Q., Wang, H. & Wang, P. Inhibition of HPV 16 E6 oncogene expression by RNA interference in vitro and in vivo. Int. J. Gynecol. Cancer 16, 743–751 (2006).
Grzelinski, M. et al. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum. Gene Ther. 17, 751–766 (2006).
Kim, W.J. et al. Cholesteryl oligoarginine delivering vascular endothelial growth factor siRNA effectively inhibits tumor growth in colon adenocarcinoma. Mol. Ther. 14, 343–350 (2006).
Song, E. et al. Antibody-mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 23, 709–717 (2005).
Takei, Y., Kadomatsu, K., Yuzawa, Y., Matsuo, S. & Muramatsu, T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 64, 3365–3370 (2004).
Minakuchi, Y. et al. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 32, e109 (2004).
McNamara, J.O. et al. Cell type–specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 24, 1005–1015 (2006).
Gurzov, E.N. & Izquierdo, M. RNA interference against Hec1 inhibits tumor growth in vivo. Gene Ther. 13, 1–7 (2005).
Pulukuri, S.M. et al. RNA interference-directed knockdown of urokinase plasminogen activator and urokinase plasminogen activator receptor inhibits prostate cancer cell invasion, survival, and tumorigenicity in vivo. J. Biol. Chem. 280, 36529–36540 (2005).
Zhang, Y. et al. Engineering mucosal RNA interference in vivo. Mol. Ther. 14, 336–342 (2006).
Shankar, P., Manjunath, N. & Lieberman, J. The prospect of silencing disease using RNA interference. J. Am. Med. Assoc. 293, 1367–1373 (2005).
Zimmermann, T.S. et al. RNAi-mediated gene silencing in non-human primates. Nature 441, 111–114 (2006).
Morrissey, D.V. et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 41, 1349–1356 (2005).
Geisbert, T.W. et al. Postexposure protection of guinea pigs against a lethal ebola virus challenge is conferred by RNA interference. J. Infect. Dis. 193, 1650–1657 (2006).
Takeshita, F. & Ochiya, T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 97, 689–696 (2006).
Pal, A. et al. Systemic delivery of RafsiRNA using cationic cardiolipin liposomes silences Raf-1 expression and inhibits tumor growth in xenograft model of human prostate cancer. Int. J. Oncol. 26, 1087–1091 (2005).
Schiffelers, R.M. et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 32, e149 (2004).
Urban-Klein, B., Werth, S., Abuharbeid, S., Czubayko, F. & Aigner, A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 12, 461–466 (2005).
Hu-Lieskovan, S., Heidel, J.D., Bartlett, D.W., Davis, M.E. & Triche, T.J. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastic Ewing's sarcoma. Cancer Res. 65, 8984–8992 (2005).
Grimm, D. et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541 (2006).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Acknowledgements
We thank J. Maraganore and B. Greene for critical reading of the manuscript, M. Duckman for graphics assistance and A. Capobianco for word processing assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors are employees of Alnylam Pharmaceuticals.
Rights and permissions
About this article
Cite this article
Bumcrot, D., Manoharan, M., Koteliansky, V. et al. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol 2, 711–719 (2006). https://doi.org/10.1038/nchembio839
Issue Date:
DOI: https://doi.org/10.1038/nchembio839
This article is cited by
-
RNA interference in the era of nucleic acid therapeutics
Nature Biotechnology (2024)
-
Nanoparticle-based theranostics in nuclear medicine
Journal of Radioanalytical and Nuclear Chemistry (2024)
-
Small interfering RNA (siRNA)-based therapeutic applications against viruses: principles, potential, and challenges
Journal of Biomedical Science (2023)
-
Endogenous dual miRNA-triggered dynamic assembly of DNA nanostructures for in-situ dual siRNA delivery
Science China Materials (2023)
-
ApoE—functionalization of nanoparticles for targeted brain delivery—a feasible method for polyplexes?
Drug Delivery and Translational Research (2023)