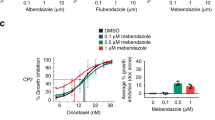
Abstract
Approved drugs are invaluable tools to study biochemical pathways, and further characterization of these compounds may lead to repurposing of single drugs or combinations. Here we describe a collection of 308 small molecules representing the diversity of structures and molecular targets of all FDA-approved chemical entities. The CeMM Library of Unique Drugs (CLOUD) covers prodrugs and active forms at pharmacologically relevant concentrations and is ideally suited for combinatorial studies. We screened pairwise combinations of CLOUD drugs for impairment of cancer cell viability and discovered a synergistic interaction between flutamide and phenprocoumon (PPC). The combination of these drugs modulates the stability of the androgen receptor (AR) and resensitizes AR-mutant prostate cancer cells to flutamide. Mechanistically, we show that the AR is a substrate for γ-carboxylation, a post-translational modification inhibited by PPC. Collectively, our data suggest that PPC could be repurposed to tackle resistance to antiandrogens in prostate cancer patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Paul, S.M. et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat. Rev. Drug Discov. 9, 203–214 (2010).
Scannell, J.W., Blanckley, A., Boldon, H. & Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11, 191–200 (2012).
Ashburn, T.T. & Thor, K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–683 (2004).
Clavel, F. & Hance, A.J. HIV drug resistance. N. Engl. J. Med. 350, 1023–1035 (2004).
Holohan, C., Van Schaeybroeck, S., Longley, D.B. & Johnston, P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13, 714–726 (2013).
Bock, C. & Lengauer, T. Managing drug resistance in cancer: lessons from HIV therapy. Nat. Rev. Cancer 12, 494–501 (2012).
Chong, C.R., Chen, X., Shi, L., Liu, J.O. & Sullivan, D.J. Jr. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat. Chem. Biol. 2, 415–416 (2006).
Huang, R. et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 3, 80ps16 (2011).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Iskar, M. et al. Drug-induced regulation of target expression. PLoS Comput. Biol. 6, e1000925 (2010).
Overington, J.P., Al-Lazikani, B. & Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 (2006).
Lipinski, C.A., Lombardo, F., Dominy, B.W. & Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 (2001).
Reiling, J.H. et al. A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc. Natl. Acad. Sci. USA 108, 11756–11765 (2011).
Winter, G.E. et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat. Chem. Biol. 10, 768–773 (2014).
Bliss, C.I. The toxicity of poisons applied jointly. Ann. Appl. Biol. 26, 585–615 (1939).
Berenbaum, M.C. What is synergy? Pharmacol. Rev. 41, 93–141 (1989).
Loewe, S. Die quantitativen Probleme der Pharmakologie. Ergeb. Physiol. 27, 48–187 (1928).
Chou, T.C. & Talalay, P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22, 27–55 (1984).
Tang, J., Wennerberg, K. & Aittokallio, T. What is synergy? The Saariselkä agreement revisited. Front. Pharmacol. 6, 181 (2015).
Bürckstümmer, T. et al. A reversible gene trap collection empowers haploid genetics in human cells. Nat. Methods 10, 965–971 (2013).
Carette, J.E. et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science 326, 1231–1235 (2009).
Dehm, S.M. & Tindall, D.J. Alternatively spliced androgen receptor variants. Endocr. Relat. Cancer 18, R183–R196 (2011).
Veldscholte, J. et al. The androgen receptor in LNCaP cells contains a mutation in the ligand binding domain which affects steroid binding characteristics and response to anti-androgens. J. Steroid Biochem. Mol. Biol. 41, 665–669 (1992).
Alimirah, F., Chen, J., Basrawala, Z., Xin, H. & Choubey, D. DU-145 and PC-3 human prostate cancer cell lines express androgen receptor: implications for the androgen receptor functions and regulation. FEBS Lett. 580, 2294–2300 (2006).
Suttie, J.W. Vitamin K-dependent carboxylase. Annu. Rev. Biochem. 54, 459–477 (1985).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Feldman, B.J. & Feldman, D. The development of androgen-independent prostate cancer. Nat. Rev. Cancer 1, 34–45 (2001).
Gaddipati, J.P. et al. Frequent detection of codon 877 mutation in the androgen receptor gene in advanced prostate cancers. Cancer Res. 54, 2861–2864 (1994).
Bhattacharyya, R.S., Krishnan, A.V., Swami, S. & Feldman, D. Fulvestrant (ICI 182,780) down-regulates androgen receptor expression and diminishes androgenic responses in LNCaP human prostate cancer cells. Mol. Cancer Ther. 5, 1539–1549 (2006).
Grad, J.M., Lyons, L.S., Robins, D.M. & Burnstein, K.L. The androgen receptor (AR) amino-terminus imposes androgen-specific regulation of AR gene expression via an exonic enhancer. Endocrinology 142, 1107–1116 (2001).
Eder, I.E. et al. Inhibition of LncaP prostate cancer cells by means of androgen receptor antisense oligonucleotides. Cancer Gene Ther. 7, 997–1007 (2000).
Liao, X., Tang, S., Thrasher, J.B., Griebling, T.L. & Li, B. Small-interfering RNA-induced androgen receptor silencing leads to apoptotic cell death in prostate cancer. Mol. Cancer Ther. 4, 505–515 (2005).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Hallgren, K.W., Zhang, D., Kinter, M., Willard, B. & Berkner, K.L. Methylation of γ-carboxylated Glu (Gla) allows detection by liquid chromatography-mass spectrometry and the identification of Gla residues in the γ-glutamyl carboxylase. J. Proteome Res. 12, 2365–2374 (2013).
Santos, R. et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017).
Taplin, M.-E. Drug insight: role of the androgen receptor in the development and progression of prostate cancer. Nat. Clin. Pract. Oncol. 4, 236–244 (2007).
de Bono, J.S. et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005 (2011).
Tran, C. et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009).
Chen, C.D. et al. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 10, 33–39 (2004).
Balbas, M.D. et al. Overcoming mutation-based resistance to anti-androgens with rational drug design. eLife 2, e00499 (2013).
Van Hemelrijck, M. et al. Cohort Profile: the National Prostate Cancer Register of Sweden and Prostate Cancer data Base Sweden 2.0. Int. J. Epidemiol. 42, 956–967 (2013).
Tagalakis, V., Tamim, H., Blostein, M., Hanley, J.A. & Kahn, S.R. Risk of prostate cancer death in long-term users of warfarin: a population-based case-control study. Cancer Causes Control 24, 1079–1085 (2013).
Tew, B.Y. et al. Vitamin K epoxide reductase regulation of androgen receptor activity. Oncotarget 8, 13818–13831 (2017).
Itsumi, M. et al. PMA induces androgen receptor downregulation and cellular apoptosis in prostate cancer cells. J. Mol. Endocrinol. 53, 31–41 (2014).
Yu, Z. et al. Galeterone prevents androgen receptor binding to chromatin and enhances degradation of mutant androgen receptor. Clin. Cancer Res. 20, 4075–4085 (2014).
Licciardello, M.P. et al. NOTCH1 activation in breast cancer confers sensitivity to inhibition of SUMOylation. Oncogene 29, 319 (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012).
Rudashevskaya, E.L. et al. A method to resolve the composition of heterogeneous affinity-purified protein complexes assembled around a common protein by chemical cross-linking, gel electrophoresis and mass spectrometry. Nat. Protoc. 8, 75–97 (2013).
Huber, M.L. et al. abFASP-MS: affinity-based filter-aided sample preparation mass spectrometry for quantitative analysis of chemically labeled protein complexes. J. Proteome Res. 13, 1147–1155 (2014).
Acknowledgements
We thank M. Iskar (EMBL) and P. Bork (EMBL) for providing us with DIPS scores, R. Schüle (Albert-Ludwigs-University Freiburg, Germany) for LAPC4 cells, and G. Winter (CeMM) and G. Superti-Furga (CeMM) for thoughtful discussions and initializing synergy screenings at CeMM. S.K. acknowledges support by a Marie Curie Career Integration Grant, the Austrian Federal Ministry of Science, Research and Economy and the National Foundation for Research, Technology, and Development and the Austrian Science Fund (FWF): F4701-B20.
Author information
Authors and Affiliations
Contributions
P.M., F.K. and S.K. designed and assembled the CLOUD. M.P.L., F.K., C.-H.L. and S.K. designed and performed the screen of the CLOUD. M.P.L., F.K., M.C. and J.M. analyzed the data from the screen. M.P.L. and A.R. designed and performed viability, RT-qPCR, western blotting, immunoprecipitation and immunofluorescence experiments. S.S. performed immunofluorescence experiments. A.R. and E.S. designed and performed gene expression and immunoprecipitation experiments. A.R. and B.B. designed and performed knockdown experiments. A.C.M. and K.L.B. designed and performed proteomics experiments. A.W., R.H. and K.K. performed and analyzed 2D gel electrophoresis experiments. T.P., M.S. and C.B. performed and analyzed RNA-seq experiments. G.D. and J.C. performed DIPS score analyses. Y.F. and P.S. analyzed patient data in PCBaSe. V.I. synthesized, provided and quality controlled chemicals. M.P.L. and S.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.P.L. and S.K. have filed a patent based on findings described in this manuscript (WO2016170102 A1). V.I. is an employee of Enamine, Ltd and may also own shares in the company.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–4 and Supplementary Figures 1–21 (PDF 29615 kb)
Supplementary Data Set 1
STEAM and CLOUD drugs. (XLS 338 kb)
Supplementary Data Set 2
Data from CLOUD combinatorial screen. (XLS 4999 kb)
Supplementary Data Set 3
Synergies and antagonisms defined by both Bliss and Loewe scores. (XLS 207 kb)
Rights and permissions
About this article
Cite this article
Licciardello, M., Ringler, A., Markt, P. et al. A combinatorial screen of the CLOUD uncovers a synergy targeting the androgen receptor. Nat Chem Biol 13, 771–778 (2017). https://doi.org/10.1038/nchembio.2382
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2382
This article is cited by
-
Augmented drug combination dataset to improve the performance of machine learning models predicting synergistic anticancer effects
Scientific Reports (2024)
-
Performance evaluation of drug synergy datasets using computational intelligence approaches
Multimedia Tools and Applications (2024)
-
A review on graph neural networks for predicting synergistic drug combinations
Artificial Intelligence Review (2024)
-
A Comprehensive Review on Deep Synergistic Drug Prediction Techniques for Cancer
Archives of Computational Methods in Engineering (2022)
-
A widespread role for SLC transmembrane transporters in resistance to cytotoxic drugs
Nature Chemical Biology (2020)