Abstract
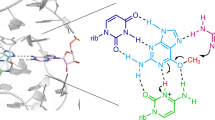
The TS ribozyme (originally called “twister sister”) is a catalytic RNA. We present a crystal structure of the ribozyme in a pre-reactive conformation. Two co-axial helical stacks are organized by a three-way junction and two tertiary contacts. Five divalent metal ions are directly coordinated to RNA ligands, making important contributions to the RNA architecture. The scissile phosphate lies in a quasihelical loop region that is organized by a network of hydrogen bonding. A divalent metal ion is directly bound to the nucleobase 5′ to the scissile phosphate, with an inner-sphere water molecule positioned to interact with the O2′ nucleophile. The rate of ribozyme cleavage correlated in a log-linear manner with divalent metal ion pKa, consistent with proton transfer in the transition state, and we propose that the bound metal ion is a likely general base for the cleavage reaction. Our data indicate that the TS ribozyme functions predominantly as a metalloenzyme.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Lilley, D.M.J. & Eckstein, F. (eds.) Ribozymes and RNA Catalysis 1–318 (The Royal Society of Chemistry, 2008).
Wilson, T.J. & Lilley, D.M.J. Biochemistry. The evolution of ribozyme chemistry. Science 323, 1436–1438 (2009).
Fedor, M.J. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry 38, 11040–11050 (1999).
McLeod, A.C. & Lilley, D.M. Efficient, pH-dependent RNA ligation by the VS ribozyme in trans. Biochemistry 43, 1118–1125 (2004).
Nahas, M.K. et al. Observation of internal cleavage and ligation reactions of a ribozyme. Nat. Struct. Mol. Biol. 11, 1107–1113 (2004).
Canny, M.D., Jucker, F.M. & Pardi, A. Efficient ligation of the Schistosoma hammerhead ribozyme. Biochemistry 46, 3826–3834 (2007).
Nakano, S., Proctor, D.J. & Bevilacqua, P.C. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry 40, 12022–12038 (2001).
Ke, A., Zhou, K., Ding, F., Cate, J.H. & Doudna, J.A. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature 429, 201–205 (2004).
Martick, M. & Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126, 309–320 (2006).
Thomas, J.M. & Perrin, D.M. Probing general acid catalysis in the hammerhead ribozyme. J. Am. Chem. Soc. 131, 1135–1143 (2009).
Klein, D.J. & Ferré-D'Amaré, A.R. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313, 1752–1756 (2006).
Cochrane, J.C., Lipchock, S.V. & Strobel, S.A. Structural investigation of the GlmS ribozyme bound to Its catalytic cofactor. Chem. Biol. 14, 97–105 (2007).
Cochrane, J.C., Lipchock, S.V., Smith, K.D. & Strobel, S.A. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry 48, 3239–3246 (2009).
Wilson, T.J. & Lilley, D.M.J. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA 17, 213–221 (2011).
Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A. & Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004).
Przybilski, R. et al. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell 17, 1877–1885 (2005).
Salehi-Ashtiani, K., Lupták, A., Litovchick, A. & Szostak, J.W. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313, 1788–1792 (2006).
Martick, M., Horan, L.H., Noller, H.F. & Scott, W.G. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 454, 899–902 (2008).
Webb, C.H., Riccitelli, N.J., Ruminski, D.J. & Lupták, A. Widespread occurrence of self-cleaving ribozymes. Science 326, 953 (2009).
Roth, A. et al. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 10, 56–60 (2014).
Weinberg, Z. et al. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 11, 606–610 (2015).
Liu, Y., Wilson, T.J., McPhee, S.A. & Lilley, D.M. Crystal structure and mechanistic investigation of the twister ribozyme. Nat. Chem. Biol. 10, 739–744 (2014).
Eiler, D., Wang, J. & Steitz, T.A. Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme. Proc. Natl. Acad. Sci. USA 111, 13028–13033 (2014).
Ren, A. et al. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat. Commun. 5, 5534 (2014).
Lilley, D.M. et al. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). A nomenclature of junctions and branchpoints in nucleic acids. Recommendations 1994. Eur. J. Biochem. 230, 1–2 (1995).
Murray, J.B., Seyhan, A.A., Walter, N.G., Burke, J.M. & Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 5, 587–595 (1998).
Das, S.R. & Piccirilli, J.A. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 1, 45–52 (2005).
Chen, J.H. et al. A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry 49, 6508–6518 (2010).
Chen, J. et al. Identification of the catalytic Mg2 ion in the hepatitis delta virus ribozyme. Biochemistry 52, 557–567 (2013).
Boots, J.L., Canny, M.D., Azimi, E. & Pardi, A. Metal ion specificities for folding and cleavage activity in the Schistosoma hammerhead ribozyme. RNA 14, 2212–2222 (2008).
Lee, T.S. et al. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J. Am. Chem. Soc. 130, 3053–3064 (2008).
Wilson, T.J., Liu, Y., Domnick, C., Kath-Schorr, S. & Lilley, D.M. The novel chemical mechanism of the twister ribozyme. J. Am. Chem. Soc. 138, 6151–6162 (2016).
Shan, S., Kravchuk, A.V., Piccirilli, J.A. & Herschlag, D. Defining the catalytic metal ion interactions in the Tetrahymena ribozyme reaction. Biochemistry 40, 5161–5171 (2001).
Stahley, M.R. & Strobel, S.A. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science 309, 1587–1590 (2005).
Forconi, M., Lee, J., Lee, J.K., Piccirilli, J.A. & Herschlag, D. Functional identification of ligands for a catalytic metal ion in group I introns. Biochemistry 47, 6883–6894 (2008).
Frederiksen, J.K., Li, N.S., Das, R., Herschlag, D. & Piccirilli, J.A. Metal-ion rescue revisited: biochemical detection of site-bound metal ions important for RNA folding. RNA 18, 1123–1141 (2012).
Crary, S.M., Kurz, J.C. & Fierke, C.A. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis ribonuclease P. RNA 8, 933–947 (2002).
Reiter, N.J. et al. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468, 784–789 (2010).
Pinard, R. et al. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 20, 6434–6442 (2001).
Wilson, T.J., Zhao, Z.Y., Maxwell, K., Kontogiannis, L. & Lilley, D.M. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 40, 2291–2302 (2001).
Bevilacqua, P.C. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry 42, 2259–2265 (2003).
Kuzmin, Y.I., Da Costa, C.P. & Fedor, M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 340, 233–251 (2004).
Han, J. & Burke, J.M. Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry 44, 7864–7870 (2005).
Klein, D.J., Been, M.D. & Ferré-D'Amaré, A.R. Essential role of an active-site guanine in glmS ribozyme catalysis. J. Am. Chem. Soc. 129, 14858–14859 (2007).
Wilson, T.J., McLeod, A.C. & Lilley, D.M.J. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 26, 2489–2500 (2007).
Wilson, T.J. et al. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc. Natl. Acad. Sci. USA 107, 11751–11756 (2010).
Kath-Schorr, S. et al. General acid-base catalysis mediated by nucleobases in the hairpin ribozyme. J. Am. Chem. Soc. 134, 16717–16724 (2012).
Feig, A.L. & Uhlenbeck, O.C. in The RNA World (eds. Gesteland, R.F., Cech, T.R. & Atkins, J.F.) 287–319 (Cold Spring Harbor Laboratory Press, 1999).
Beaucage, S.L. & Caruthers, M.H. Deoxynucleoside phosphoramidites - a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedr. Lett. 22, 1859–1862 (1981).
Hakimelahi, G.H., Proba, Z.A. & Ogilvie, K.K. High yield selective 3′-silylation of ribonucleosides. Tetrahedr. Lett. 22, 5243–5246 (1981).
Perreault, J.-P., Wu, T.F., Cousineau, B., Ogilvie, K.K. & Cedergren, R. Mixed deoxyribo- and ribo-oligonucleotides with catalytic activity. Nature 344, 565–567 (1990).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Acknowledgements
We thank S. Ashraf for expert synthesis of RNA, the CRUK for program support A18604 (to D.M.J.L.), the Wellcome Trust for the in-house diffractometer and ESRF for synchrotron beam time.
Author information
Authors and Affiliations
Contributions
Y.L. performed crystallography, T.J.W. performed mechanistic investigations and Y.L., T.J.W. and D.M.J.L. designed the research, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–12 (PDF 10052 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Wilson, T. & Lilley, D. The structure of a nucleolytic ribozyme that employs a catalytic metal ion. Nat Chem Biol 13, 508–513 (2017). https://doi.org/10.1038/nchembio.2333
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2333
This article is cited by
-
Theoretical Studies of the Self Cleavage Pistol Ribozyme Mechanism
Topics in Catalysis (2022)
-
LCS-TA to identify similar fragments in RNA 3D structures
BMC Bioinformatics (2017)
-
Structure-based insights into self-cleavage by a four-way junctional twister-sister ribozyme
Nature Communications (2017)
-
Nanopore electric snapshots of an RNA tertiary folding pathway
Nature Communications (2017)