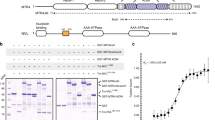
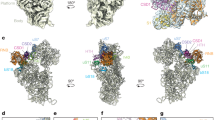
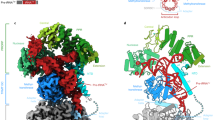
Abstract
The exosome is a large molecular machine involved in RNA degradation and processing. Here we address how the trimeric Rrp4 cap enhances the activity of the archaeal enzyme complex. Using methyl-TROSY NMR methods we identified a 50-Å long RNA binding path on each Rrp4 protomer. We show that the Rrp4 cap can thus simultaneously recruit three substrates, one of which is degraded in the core while the others are positioned for subsequent degradation rounds. The local interaction energy between the substrate and the Rrp4–exosome increases from the periphery of the complex toward the active sites. Notably, the intrinsic interaction strength between the cap and the substrate is weakened as soon as substrates enter the catalytic barrel, which provides a means to reduce friction during substrate movements toward the active sites. Our data thus reveal a sophisticated exosome–substrate interaction mechanism that enables efficient RNA degradation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M. & Tollervey, D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91, 457–466 (1997).
Bousquet-Antonelli, C., Presutti, C. & Tollervey, D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102, 765–775 (2000).
Mitchell, P., Petfalski, E. & Tollervey, D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 10, 502–513 (1996).
van Hoof, A., Frischmeyer, P.A., Dietz, H.C. & Parker, R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264 (2002).
Dziembowski, A., Lorentzen, E., Conti, E. & Séraphin, B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 14, 15–22 (2007).
Evguenieva-Hackenberg, E., Walter, P., Hochleitner, E., Lottspeich, F. & Klug, G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 4, 889–893 (2003).
Walter, P. et al. Characterization of native and reconstituted exosome complexes from the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 62, 1076–1089 (2006).
Roppelt, V., Klug, G. & Evguenieva-Hackenberg, E. The evolutionarily conserved subunits Rrp4 and Csl4 confer different substrate specificities to the archaeal exosome. FEBS Lett. 584, 2931–2936 (2010).
Wasmuth, E.V., Januszyk, K. & Lima, C.D. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 511, 435–439 (2014).
Liu, Q., Greimann, J.C. & Lima, C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127, 1223–1237 (2006).
Makino, D.L., Baumgärtner, M. & Conti, E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 495, 70–75 (2013).
Lorentzen, E. et al. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 12, 575–581 (2005).
Navarro, M.V., Oliveira, C.C., Zanchin, N.I. & Guimarães, B.G. Insights into the mechanism of progressive RNA degradation by the archaeal exosome. J. Biol. Chem. 283, 14120–14131 (2008).
Lorentzen, E. & Conti, E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol. Cell 20, 473–481 (2005).
Lorentzen, E., Dziembowski, A., Lindner, D., Seraphin, B. & Conti, E. RNA channelling by the archaeal exosome. EMBO Rep. 8, 470–476 (2007).
Büttner, K., Wenig, K. & Hopfner, K.P. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell 20, 461–471 (2005).
Hartung, S., Niederberger, T., Hartung, M., Tresch, A. & Hopfner, K.P. Quantitative analysis of processive RNA degradation by the archaeal RNA exosome. Nucleic Acids Res. 38, 5166–5176 (2010).
Audin, M.J., Wurm, J.P., Cvetkovic, M.A. & Sprangers, R. The oligomeric architecture of the archaeal exosome is important for processive and efficient RNA degradation. Nucleic Acids Res. 44, 2962–2973 (2016).
Koonin, E.V., Wolf, Y.I. & Aravind, L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 11, 240–252 (2001).
Niederberger, T., Hartung, S., Hopfner, K.P. & Tresch, A. Processive RNA decay by the exosome: merits of a quantitative Bayesian sampling approach. RNA Biol. 8, 55–60 (2011).
Wiesner, S. & Sprangers, R. Methyl groups as NMR probes for biomolecular interactions. Curr. Opin. Struct. Biol. 35, 60–67 (2015).
Kerfah, R., Plevin, M.J., Sounier, R., Gans, P. & Boisbouvier, J. Methyl-specific isotopic labeling: a molecular tool box for solution NMR studies of large proteins. Curr. Opin. Struct. Biol. 32, 113–122 (2015).
Gardner, K.H. & Kay, L.E. Production and incorporation of 15N, 13C, 2H (1H-δ1 methyl) isoleucine into proteins for multidimensional NMR studies. J. Am. Chem. Soc. 119, 7599–7600 (1997).
Tugarinov, V., Hwang, P.M., Ollerenshaw, J.E. & Kay, L.E. Cross-correlated relaxation enhanced 1H–13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 125, 10420–10428 (2003).
Audin, M.J. et al. The archaeal exosome: identification and quantification of site-specific motions that correlate with cap and RNA binding. Angew. Chem. Int. Edn Engl. 52, 8312–8316 (2013).
Gelis, I. et al. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131, 756–769 (2007).
Rosenzweig, R., Moradi, S., Zarrine-Afsar, A., Glover, J.R. & Kay, L.E. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 339, 1080–1083 (2013).
Sprangers, R. & Kay, L.E. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445, 618–622 (2007).
Mari, S. et al. Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure 22, 1639–1649 (2014).
Stoffregen, M.C., Schwer, M.M., Renschler, F.A. & Wiesner, S. Methionine scanning as an NMR tool for detecting and analyzing biomolecular interaction surfaces. Structure 20, 573–581 (2012).
Rosenzweig, R. & Kay, L.E. Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu. Rev. Biochem. 83, 291–315 (2014).
Sprangers, R., Gribun, A., Hwang, P.M., Houry, W.A. & Kay, L.E. Quantitative NMR spectroscopy of supramolecular complexes: dynamic side pores in ClpP are important for product release. Proc. Natl. Acad. Sci. USA 102, 16678–16683 (2005).
Amero, C. et al. A systematic mutagenesis-driven strategy for site-resolved NMR studies of supramolecular assemblies. J. Biomol. NMR 50, 229–236 (2011).
Hollingworth, D. et al. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 40, 6873–6886 (2012).
Chekanova, J.A., Dutko, J.A., Mian, I.S. & Belostotsky, D.A. Arabidopsis thaliana exosome subunit AtRrp4p is a hydrolytic 3→5′ exonuclease containing S1 and KH RNA-binding domains. Nucleic Acids Res. 30, 695–700 (2002).
Holm, L. & Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Jencks, W.P. On the attribution and additivity of binding energies. Proc. Natl. Acad. Sci. USA 78, 4046–4050 (1981).
Searle, M.S. & Williams, D.H. On the stability of nucleic acid structures in solution: enthalpy-entropy compensations, internal rotations and reversibility. Nucleic Acids Res. 21, 2051–2056 (1993).
Waudby, C.A., Ramos, A., Cabrita, L.D. & Christodoulou, J. Two-dimensional NMR lineshape analysis. Sci. Rep. 6, 24826 (2016).
Kovrigin, E.L. NMR line shapes and multi-state binding equilibria. J. Biomol. NMR 53, 257–270 (2012).
Bain, A.D., Rex, D.M. & Smith, R.N. Fitting dynamic NMR lineshapes. Magn. Reson. Chem. 39, 122–126 (2001).
Makino, D.L. et al. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 524, 54–58 (2015).
Vuković, L., Chipot, C., Makino, D.L., Conti, E. & Schulten, K. Molecular mechanism of processive 3′ to 5′ RNA translocation in the activesSubunit of the RNA exosome complex. J. Am. Chem. Soc. 138, 4069–4078 (2016).
Zhou, H.X. & Gilson, M.K. Theory of free energy and entropy in noncovalent binding. Chem. Rev. 109, 4092–4107 (2009).
Butner, K.A. & Kirschner, M.W. Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 115, 717–730 (1991).
Saio, T., Guan, X., Rossi, P., Economou, A. & Kalodimos, C.G. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 344, 1250494 (2014).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Pervushin, K., Riek, R., Wider, G. & Wüthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 94, 12366–12371 (1997).
Ramos, A. & Varani, G. A new method to detect long-range protein-RNA contacts: NMR detection of electron-proton relaxation induced by nitroxide spin-labeled RNA. J. Am. Chem. Soc. 120, 10992–10993 (1998).
Johnson, P.E., Tomme, P., Joshi, M.D. & McIntosh, L.P. Interaction of soluble cellooligosaccharides with the N-terminal cellulose-binding domain of Cellulomonas fimi CenC 2. NMR and ultraviolet absorption spectroscopy. Biochemistry 35, 13895–13906 (1996).
Acknowledgements
We acknowledge all members of R.S.'s laboratory for discussions. We thank S. Wiesner for suggestions regarding the methionine scanning experiments, I. Holdermann and J. Petters for excellent technical assistance and V. Truffault for maintenance of the NMR infrastructure. M.A.C. and S.S. acknowledge funding from the International Max Planck Research School “From Molecules to Organisms”. This work was supported by the Max Planck Society and the European Research Council under the European Union's Seventh Framework Programme (FP7/2007–2013), ERC grant agreement 616052 (R.S.).
Author information
Authors and Affiliations
Contributions
M.A.C. performed and analyzed NMR, binding and degradation experiments. J.P.W. assisted with the binding experiments, M.J.A. assisted with the degradation experiments and S.S. performed NMR assignment experiments. R.S. conceived the project, analyzed data and wrote the manuscript. All authors commented on the data, the analysis and the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–17 (PDF 13162 kb)
Rights and permissions
About this article
Cite this article
Cvetkovic, M., Wurm, J., Audin, M. et al. The Rrp4–exosome complex recruits and channels substrate RNA by a unique mechanism. Nat Chem Biol 13, 522–528 (2017). https://doi.org/10.1038/nchembio.2328
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2328