Abstract
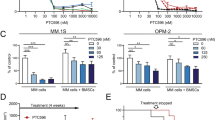
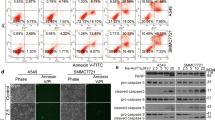
The proteasome is a vital cellular machine that maintains protein homeostasis, which is of particular importance in multiple myeloma and possibly other cancers. Targeting of proteasome 20S peptidase activity with bortezomib and carfilzomib has been widely used to treat myeloma. However, not all patients respond to these compounds, and those who do eventually suffer relapse. Therefore, there is an urgent and unmet need to develop new drugs that target proteostasis through different mechanisms. We identified quinoline-8-thiol (8TQ) as a first-in-class inhibitor of the proteasome 19S subunit Rpn11. A derivative of 8TQ, capzimin, shows >5-fold selectivity for Rpn11 over the related JAMM proteases and >2 logs selectivity over several other metalloenzymes. Capzimin stabilized proteasome substrates, induced an unfolded protein response, and blocked proliferation of cancer cells, including those resistant to bortezomib. Proteomic analysis revealed that capzimin stabilized a subset of polyubiquitinated substrates. Identification of capzimin offers an alternative path to develop proteasome inhibitors for cancer therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009).
Dimopoulos, M.A., Richardson, P.G., Moreau, P. & Anderson, K.C. Current treatment landscape for relapsed and/or refractory multiple myeloma. Nat. Rev. Clin. Oncol. 12, 42–54 (2015).
Verma, R. et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 (2002).
Yao, T. & Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 (2002).
Ambroggio, X.I., Rees, D.C. & Deshaies, R.J. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2, E2 (2004).
Tran, H.J., Allen, M.D., Löwe, J. & Bycroft, M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 42, 11460–11465 (2003).
Cope, G.A. et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298, 608–611 (2002).
Zhu, P. et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell 27, 609–621 (2007).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007).
McCullough, J., Clague, M.J. & Urbé, S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166, 487–492 (2004).
Gallery, M. et al. The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Mol. Cancer Ther. 6, 262–268 (2007).
Bivona, T.G. et al. FAS and NF-κB signalling modulate dependence of lung cancers on mutant EGFR. Nature 471, 523–526 (2011).
Buckley, S.M. et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 11, 783–798 (2012).
Perez, C. et al. Discovery of an inhibitor of the proteasome subunit Rpn11. J. Med. Chem. (10.1021/acs.jmedchem.6b01379. published online 13 February 2017).
Dantuma, N.P., Lindsten, K., Glas, R., Jellne, M. & Masucci, M.G. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18, 538–543 (2000).
Rouffet, M. & Cohen, S.M. Emerging trends in metalloprotein inhibition. Dalton Trans. 40, 3445–3454 (2011).
Jacobsen, F.E., Lewis, J.A. & Cohen, S.M. The design of inhibitors for medicinally relevant metalloproteins. ChemMedChem 2, 152–171 (2007).
Dambacher, C.M., Worden, E.J., Herzik, M.A., Martin, A. & Lander, G.C. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife 5, e13027 (2016).
Worden, E.J., Padovani, C. & Martin, A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat. Struct. Mol. Biol. 21, 220–227 (2014).
Pathare, G.R. et al. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc. Natl. Acad. Sci. USA 111, 2984–2989 (2014).
Henderson, A., Erales, J., Hoyt, M.A. & Coffino, P. Dependence of proteasome processing rate on substrate unfolding. J. Biol. Chem. 286, 17495–17502 (2011).
Carter, K.P., Young, A.M. & Palmer, A.E. Fluorescent sensors for measuring metal ions in living systems. Chem. Rev. 114, 4564–4601 (2014).
Zeqiraj, E. et al. Higher-order assembly of BRCC36-KIAA0157 is required for DUB activity and biological function. Mol. Cell 59, 970–983 (2015).
Sato, Y. et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 (2008).
Chou, T.F. & Deshaies, R.J. Quantitative cell-based protein degradation assays to identify and classify drugs that target the ubiquitin-proteasome system. J. Biol. Chem. 286, 16546–16554 (2011).
Johnston, J.A., Ward, C.L. & Kopito, R.R. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898 (1998).
Hao, R. et al. Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains. Mol. Cell 51, 819–828 (2013).
Anderson, D.J. et al. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell 28, 653–665 (2015).
Chou, T.F. et al. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc. Natl. Acad. Sci. USA 108, 4834–4839 (2011).
Wacker, S.A., Houghtaling, B.R., Elemento, O. & Kapoor, T.M. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat. Chem. Biol. 8, 235–237 (2012).
Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 6, 813–823 (2006).
Xu, Y. & Price, B.D. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle 10, 261–267 (2011).
Chan, J.Y. et al. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 17, 1779–1787 (1998).
Steffen, J., Seeger, M., Koch, A. & Krüger, E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 40, 147–158 (2010).
Radhakrishnan, S.K. et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38, 17–28 (2010).
Lee, M.J., Lee, B.H., Hanna, J., King, R.W. & Finley, D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell. Proteomics 10, R110.003871 (2011).
Lv, M. et al. Angiomotin promotes breast cancer cell proliferation and invasion. Oncol. Rep. 33, 1938–1946 (2015).
Bush, K.T., Goldberg, A.L. & Nigam, S.K. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J. Biol. Chem. 272, 9086–9092 (1997).
Holland, A.J. & Cleveland, D.W. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10, 478–487 (2009).
Deshaies, R.J. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 12, 94 (2014).
Johnson, D.E. The ubiquitin-proteasome system: opportunities for therapeutic intervention in solid tumors. Endocr. Relat. Cancer 22, T1–T17 (2015).
Trader, D.J., Simanski, S. & Kodadek, T. A reversible and highly selective inhibitor of the proteasomal ubiquitin receptor rpn13 is toxic to multiple myeloma cells. J. Am. Chem. Soc. 137, 6312–6319 (2015).
Lim, H.S., Archer, C.T. & Kodadek, T. Identification of a peptoid inhibitor of the proteasome 19S regulatory particle. J. Am. Chem. Soc. 129, 7750–7751 (2007).
D'Arcy, P. et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 17, 1636–1640 (2011).
Anchoori, R.K. et al. A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell 24, 791–805 (2013).
Al-Shami, A. et al. Regulators of the proteasome pathway, Uch37 and Rpn13, play distinct roles in mouse development. PLoS One 5, e13654 (2010).
Radhakrishnan, S.K., den Besten, W. & Deshaies, R.J. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. eLife 3, e01856 (2014).
Ong, C.K., Lirk, P., Tan, C.H. & Seymour, R.A. An evidence-based update on nonsteroidal anti-inflammatory drugs. Clin. Med. Res. 5, 19–34 (2007).
Marks, P.A. & Breslow, R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 25, 84–90 (2007).
Borg-Neczak, K. & Tjälve, H. Effect of 8-hydroxy-, 8-mercapto- and 5-chloro-7-iodo-8-hydroxy-quinoline on the uptake and distribution of nickel in mice. Pharmacol. Toxicol. 74, 185–192 (1994).
Duda, D.M. et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008).
Puerta, D.T. et al. Heterocyclic zinc-binding groups for use in next-generation matrix metalloproteinase inhibitors: potency, toxicity, and reactivity. J. Biol. Inorg. Chem. 11, 131–138 (2006).
Martin, D.P., Hann, Z.S. & Cohen, S.M. Metalloprotein-inhibitor binding: human carbonic anhydrase II as a model for probing metal-ligand interactions in a metalloprotein active site. Inorg. Chem. 52, 12207–12215 (2013).
Hess, S., van Beek, J. & Pannell, L.K. Acid hydrolysis of silk fibroins and determination of the enrichment of isotopically labeled amino acids using precolumn derivatization and high-performance liquid chromatography-electrospray ionization-mass spectrometry. Anal. Biochem. 311, 19–26 (2002).
Sung, M.K. et al. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. eLife 5, e19105 (2016).
Udeshi, N.D. et al. Refined preparation and use of anti-diglycine remnant (K-ɛ-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 (2013).
Porras-Yakushi, T.R., Sweredoski, M.J. & Hess, S. ETD outperforms CID and HCD in the analysis of the ubiquitylated proteome. J. Am. Soc. Mass Spectrom. 26, 1580–1587 (2015).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Acknowledgements
We thank A. Martin (University of California, Berkeley) for the plasmid that expresses the Rpn11–Rpn8 dimer, E. Zeqiraj (University of Leeds) for providing purified BRISC complex, P. Coffino (The Rockefeller University) for providing the Rpn10-I27V13Pext plasmid, T.M. Kapoor (The Rockefeller University) for providing the RPE1 WT and BTZ-resistant cell lines, National Cancer Institute (Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis) for screening capzimin against 60 human cancer cell lines (http://dtp.cancer.gov), H. Park for testing the cell lines for mycoplasma contamination and Y. Zhang for advice on the competition studies. This work was supported by grants from the Caltech Gates Grubstake Fund and Amgen to R.J.D. and from the NIH (CA164803) to R.J.D. and S.M.C., T.Y. and S.H. were supported by the Gordon and Betty Moore Gordon Foundation, through Grant GBMF775, the Beckman Institute, and the NIH through Grant 1S10RR029594-01A1. R.J.D. is an Investigator of and was supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
J.L. designed, executed, and interpreted all experiments with capzimin, screened the MBP library, and drafted the manuscript. T.Y. executed and interpreted mass spectrometry proteomic analysis with capzimin. F.P. was responsible for the initial development of Rpn11 HTS substrate and fluorescence polarization assay and design and execution of Rpn11 HTS. A.L.M. triaged Rpn11 HTS hits. C.P. executed HDAC6, MMP, hCAII, and GLO1 assay and synthesis of 3021 and 3027 (capzimin). Y.M. synthesized 3027 (capzimin). S.C., G.M., B.B., E. Sergienko, and I.P. developed the HTS assay. K.N., S.V., E. Suyama, and L.H.S. performed the HTS assay. I.P., A.B.P., E. Sergienko, and T.D.Y.C. evaluated and selected hits. K.P.C. designed, executed, and interpreted experiments with zinc reporter. A.E.P. designed and interpreted experiments with zinc reporter. S.H. designed, interpreted and oversaw the mass spectrometric studies with capzimin and revised the manuscript. S.M.C. designed and oversaw capzimin syntheses as well as aided in interpretation of metalloenzyme inhibition studies. R.J.D. designed, interpreted, and oversaw the experiments for the entire study and drafted the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.J.D. is a founder, shareholder, consultant, and member of the Scientific Advisory Board of Cleave Biosciences, which is engaged in discovery and development of drugs that target enzymes involved in ubiquitin-dependent protein degradation. S.M.C. is a co-founder, has an equity interest, and receives income as member of the Scientific Advisory Board for Cleave Biosciences and is a co-founder, has an equity interest, and is a member of the Scientific Advisory Board for Forge Therapeutics. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–5 and Supplementary Figures 1–21 (PDF 2331 kb)
Supplementary Note
Synthesis of 3021 and 3027 (CZM) (PDF 75 kb)
Dataset 1
GlyGly Table information (XLSX 2008 kb)
Dataset 2
Protein Groups with information (XLSX 1594 kb)
Rights and permissions
About this article
Cite this article
Li, J., Yakushi, T., Parlati, F. et al. Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat Chem Biol 13, 486–493 (2017). https://doi.org/10.1038/nchembio.2326
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2326
This article is cited by
-
Depletion of proteasome subunit PSMD1 induces cancer cell death via protein ubiquitination and DNA damage, irrespective of p53 status
Scientific Reports (2024)
-
The function and mechanism of PSMD14 in promoting progression and resistance to anlotinib in osteosarcoma
Cancer Cell International (2023)
-
The prognostic role of PSMD14 in head and neck squamous cell carcinoma
Journal of Cancer Research and Clinical Oncology (2023)
-
The emerging role of deubiquitylating enzymes as therapeutic targets in cancer metabolism
Cancer Cell International (2022)
-
A drug repurposing strategy for overcoming human multiple myeloma resistance to standard-of-care treatment
Cell Death & Disease (2022)