Abstract
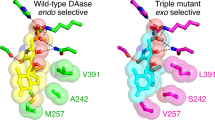
The asymmetric dehydration of alcohols is an important process for the direct synthesis of alkenes. We report the structure and substrate specificity of the bifunctional linalool dehydratase isomerase (LinD) from the bacterium Castellaniella defragrans that catalyzes in nature the hydration of β-myrcene to linalool and the subsequent isomerization to geraniol. Enzymatic kinetic resolutions of truncated and elongated aromatic and aliphatic tertiary alcohols (C5–C15) that contain a specific signature motif demonstrate the broad substrate specificity of LinD. The three-dimensional structure of LinD from Castellaniella defragrans revealed a pentamer with active sites at the protomer interfaces. Furthermore, the structure of LinD in complex with the product geraniol provides initial mechanistic insights into this bifunctional enzyme. Site-directed mutagenesis confirmed active site amino acid residues essential for its dehydration and isomerization activity. These structural and mechanistic insights facilitate the development of hydrating catalysts, enriching the toolbox for novel bond-forming biocatalysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Frija, L.M.T. & Afonso, C.A.M. Amberlyst-15: a reusable heterogeneous catalyst for the dehydration of tertiary alcohols. Tetrahedron 68, 7414–7421 (2012).
Alvarez-Manzaneda, E.J. et al. Triphenylphosphine-iodine: an efficient reagent for the regioselective dehydration of tertiary alcohols. Tetrahedr. Lett. 45, 4453–4455 (2004).
Raju, S., Moret, M.E. & Klein Gebbink, R.J.M. Rhenium-catalyzed dehydration and deoxydehydration of alcohols and polyols: opportunities for the formation of olefins from biomass. ACS Catal. 5, 281–300 (2015).
Kantam, M.L., Santhi, P.K. & Siddiqui, M.F. Montmorillonite-catalyzed dehydration of tertiary alcohols to olefins. Tetrahedr. Lett. 34, 1185–1186 (1993).
Kantam, M.L., Prasad, A.D. & Santhi, P.L. Molybdenum-catalyzed dehydration of tertiary alcohols to olefins. Synth. Commun. 23, 45–48 (1993).
Posner, G.H. et al. Boron trifluoride etherate promotes fast, mild, clean and regioselective dehydration of tertiary alcohols. Tetrahedr. Lett. 32, 6489–6492 (1991).
Zhang, X. et al. Low temperature dehydrations of non-activated alcohols via halide catalysis. Org. Chem. Front. 3, 701–708 (2016).
Manabe, K., Iimura, S., Sun, X.-M. & Kobayashi, S. Dehydration reactions in water. Brønsted acid-surfactant-combined catalyst for ester, ether, thioether, and dithioacetal formation in water. J. Am. Chem. Soc. 124, 11971–11978 (2002).
Harmer, M.A. & Sun, Q. Solid acid catalysis using ion-exchange resins. Appl. Catal. A Gen. 221, 45–62 (2001).
Resch, V. & Hanefeld, U. The selective addition of water. Catal. Sci. Technol. 5, 1385–1399 (2015).
Jin, J. & Hanefeld, U. The selective addition of water to C=C bonds; enzymes are the best chemists. Chem. Commun. (Camb.) 47, 2502–2510 (2011).
Tong, I.T., Liao, H.H. & Cameron, D.C. 1,3-Propanediol production by Escherichia coli expressing genes from the Klebsiella pneumoniae dha regulon. Appl. Environ. Microbiol. 57, 3541–3546 (1991).
Biebl, H., Menzel, K., Zeng, A.-P. & Deckwer, W.-D. Microbial production of 1,3-propanediol. Appl. Microbiol. Biotechnol. 52, 289–297 (1999).
Jiang, W., Wang, S., Wang, Y. & Fang, B. Key enzymes catalyzing glycerol to 1,3-propanediol. Biotechnol. Biofuels 9, 57 (2016).
Liu, J.Z., Xu, W., Chistoserdov, A. & Bajpai, R.K. Glycerol dehydratases: biochemical structures, catalytic mechanisms, and industrial applications in 1,3-propanediol production by naturally occurring and genetically engineered bacterial strains. Appl. Biochem. Biotechnol. 179, 1073–1100 (2016).
Volkov, A. et al. Myosin cross-reactive antigen of Streptococcus pyogenes M49 encodes a fatty acid double bond hydratase that plays a role in oleic acid detoxification and bacterial virulence. J. Biol. Chem. 285, 10353–10361 (2010).
Turbek, C.S., Smith, D.A. & Schardl, C.L. An extracellular enzyme from Fusarium solani f. sp. phaseoli which catalyses hydration of the isoflavonoid phytoalexin, phaseollidin. FEMS Microbiol. Lett. 73, 187–190 (1992).
Wuensch, C. et al. Asymmetric enzymatic hydration of hydroxystyrene derivatives. Angew. Chem. Int. Ed. Engl. 52, 2293–2297 (2013).
Brodkorb, D., Gottschall, M., Marmulla, R., Lüddeke, F. & Harder, J. Linalool dehydratase-isomerase, a bifunctional enzyme in the anaerobic degradation of monoterpenes. J. Biol. Chem. 285, 30436–30442 (2010).
Lüddeke, F. et al. Geraniol and geranial dehydrogenases induced in anaerobic monoterpene degradation by Castellaniella defragrans. Appl. Environ. Microbiol. 78, 2128–2136 (2012).
Lüddeke, F., Dikfidan, A. & Harder, J. Physiology of deletion mutants in the anaerobic β-myrcene degradation pathway in Castellaniella defragrans. BMC Microbiol. 12, 192 (2012).
Foss, S., Heyen, U. & Harder, J. Alcaligenes defragrans sp. nov., description of four strains isolated on alkenoic monoterpenes ((+)-menthene, alpha-pinene, 2-carene, and alpha-phellandrene) and nitrate. Syst. Appl. Microbiol. 21, 237–244 (1998).
Heyen, U. & Harder, J. Geranic acid formation, an initial reaction of anaerobic monoterpene metabolism in denitrifying Alcaligenes defragrans. Appl. Environ. Microbiol. 66, 3004–3009 (2000).
Marmulla, R., Šafaric, B., Markert, S., Schweder, T. & Harder, J. Linalool isomerase, a membrane-anchored enzyme in the anaerobic monoterpene degradation in Thauera linaloolentis 47Lol. BMC Biochem. 17, 1–11 (2016).
Weidenweber, S., Marmulla, R., Ermler, U. & Harder, J. X-ray structure of linalool dehydratase/isomerase from Castellaniella defragrans reveals enzymatic alkene synthesis. FEBS Lett. 590, 1375–1383 (2016).
Lüddeke, F. & Harder, J. Enantiospecific (S)-(+)-linalool formation from β-myrcene by linalool dehydratase-isomerase. Z. Naturforsch. C 66, 409–412 (2011).
Holm, L. & Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Itoh, T., Ochiai, A., Mikami, B., Hashimoto, W. & Murata, K. Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate. Biochem. Biophys. Res. Commun. 347, 1021–1029 (2006).
Fujiwara, T. et al. Crystal structure of Ruminococcus albus cellobiose 2-epimerase: structural insights into epimerization of unmodified sugar. FEBS Lett. 587, 840–846 (2013).
Oldfield, E. & Lin, F.Y. Terpene biosynthesis: modularity rules. Angew. Chem. Int. Ed. Engl. 51, 1124–1137 (2012).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Lakshminarasimhan, M., Madzelan, P., Nan, R., Milkovic, N.M. & Wilson, M.A. Evolution of new enzymatic function by structural modulation of cysteine reactivity in Pseudomonas fluorescens isocyanide hydratase. J. Biol. Chem. 285, 29651–29661 (2010).
Gao, J., Liao, J. & Yang, G.Y. CAAX-box protein, prenylation process and carcinogenesis. Am. J. Transl. Res. 1, 312–325 (2009).
Levine, R.L., Mosoni, L., Berlett, B.S. & Stadtman, E.R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA 93, 15036–15040 (1996).
Stadtman, E.R., Moskovitz, J., Berlett, B.S. & Levine, R.L. Cyclic oxidation and reduction of protein methionine residues is an important antioxidant mechanism. Mol. Cell. Biochem. 234-235, 3–9 (2002).
Marliere, P., Delcourt, M. & Mazaleyrat, S. Alkenol dehydratase variants. WO patent 2014184345 A1 (2014).
Overman, L.E. A general method for the synthesis of amines by the rearrangement of allylic trichloroacetimidates. 1,3-Transposition of alcohol and amine functions. J. Am. Chem. Soc. 98, 2901–2910 (1976).
Willimann, L. & Schinz, H. Über 7-Methyl-octadien-(2,6)-ol-(1), ein 'Apogeraniol'. Helv. Chim. Acta 35, 2401–2406 (1952).
Schwartz, B.D. et al. Towards the total synthesis of vibsanin E, 15-O-methylcyclovibsanin B, 3-hydroxyvibsanin E, furanovibsanin A, and 3-O-methylfuranovibsanin A. Eur. J. Org. Chem. 2006, 3181–3192 (2006).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P.R. & Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Winter, G. xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 (2010).
Sheldrick, G.M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 (2010).
Cowtan, K. Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 (2010).
Cowtan, K. Fitting molecular fragments into electron density. Acta Crystallogr. D Biol. Crystallogr. 64, 83–89 (2008).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Acknowledgements
This project has received funding from the European Union's Horizon 2020 research and innovation programme (EmPowerPutida) under grant agreement no. 635536 (to B.H.). This work was supported by the Diamond Light Source for access to beamlines I02 and I04 under proposed number mx-9948 (to G.G.).
Author information
Authors and Affiliations
Contributions
S.P., H.T.H., H.M., M.O., J.P.T. and G.G. conceived and designed the crystallization experiments and performed 3D structure determination. S.R., R.J.H., R.S., M.A.N., E.C.R. S.V.D. and S.J.C. created and characterized all of the LinD mutants used. C.G., M.-P.F. and B.M.N. performed biotransformations and contributed to data analysis. B.M.N., G.G. and B.H. wrote the manuscript. All authors revised and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–4. (PDF 828 kb)
Supplementary Note
Synthetic Procedures. (PDF 640 kb)
Rights and permissions
About this article
Cite this article
Nestl, B., Geinitz, C., Popa, S. et al. Structural and functional insights into asymmetric enzymatic dehydration of alkenols. Nat Chem Biol 13, 275–281 (2017). https://doi.org/10.1038/nchembio.2271
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2271
This article is cited by
-
Direct 1,3-butadiene biosynthesis in Escherichia coli via a tailored ferulic acid decarboxylase mutant
Nature Communications (2021)
-
A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nature Catalysis (2019)
-
Extending the application of biocatalysis to meet the challenges of drug development
Nature Reviews Chemistry (2018)
-
On the current role of hydratases in biocatalysis
Applied Microbiology and Biotechnology (2018)