Abstract
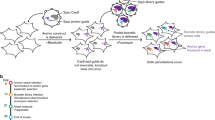
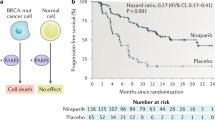
In model organisms, classical genetic screening via random mutagenesis provides key insights into the molecular bases of genetic interactions, helping to define synthetic lethality, synthetic viability and drug-resistance mechanisms. The limited genetic tractability of diploid mammalian cells, however, precludes this approach. Here, we demonstrate the feasibility of classical genetic screening in mammalian systems by using haploid cells, chemical mutagenesis and next-generation sequencing, providing a new tool to explore mammalian genetic interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Forsburg, S.L. Nat. Rev. Genet. 2, 659–668 (2001).
St Johnston, D. Nat. Rev. Genet. 3, 176–188 (2002).
Boutros, M. & Ahringer, J. Nat. Rev. Genet. 9, 554–566 (2008).
Carette, J.E. et al. Science 326, 1231–1235 (2009).
Koike-Yusa, H., Li, Y., Tan, E.-P., Velasco-Herrera, Mdel.C. & Yusa, K. Nat. Biotechnol. 32, 267–273 (2014).
Shalem, O. et al. Science 343, 84–87 (2014).
Rolef Ben-Shahar, T. et al. Science 321, 563–566 (2008).
Leeb, M. & Wutz, A. Nature 479, 131–134 (2011).
Munroe, R. & Schimenti, J. Methods Mol. Biol. 530, 131–138 (2009).
Lepage, G.A. & Jones, M. Cancer Res. 21, 642–649 (1961).
Swann, P.F. et al. Science 273, 1109–1111 (1996).
Guo, G., Wang, W. & Bradley, A. Nature 429, 891–895 (2004).
Elling, U. et al. Cell Stem Cell 9, 563–574 (2011).
Jinnah, H.A., De Gregorio, L., Harris, J.C., Nyhan, W.L. & O'Neill, J.P. Mutat. Res. 463, 309–326 (2000).
Jiricny, J. Cold Spring Harb. Perspect. Biol. 5, a012633 (2013).
Loughery, J.E.P. et al. Hum. Mol. Genet. 20, 3241–3255 (2011).
Kasap, C., Elemento, O. & Kapoor, T.M. Nat. Chem. Biol. 10, 626–628 (2014).
Smurnyy, Y. et al. Nat. Chem. Biol. 10, 623–625 (2014).
Blomen, V.A. et al. Science 350, 1092–1096 (2015).
Sagi, I. et al. Nature 532, 107–111 (2016).
Li, H. et al. Bioinformatics 25, 2078–2079 (2009).
Li, H. & Durbin, R. Bioinformatics 25, 1754–1760 (2009).
McLaren, W. et al. Bioinformatics 26, 2069–2070 (2010).
Danecek, P. et al. Bioinformatics 27, 2156–2158 (2011).
Keane, T.M. et al. Nature 477, 289–294 (2011).
Narzisi, G. et al. Nat. Methods 11, 1033–1036 (2014).
Hong, Z. et al. J. Cell Sci. 121, 3146–3154 (2008).
Murakami, K. et al. Nature 529, 403–407 (2016).
Yusa, K., Zhou, L., Li, M.A., Bradley, A. & Craig, N.L. Proc. Natl. Acad. Sci. USA 108, 1531–1536 (2011).
Choi, Y. & Chan, A.P. Bioinformatics 31, 2745–2747 (2015).
Kumar, P., Henikoff, S. & Ng, P.C. Nat. Protoc. 4, 1073–1081 (2009).
Chiang, T.-W.W., le Sage, C., Larrieu, D., Demir, M. & Jackson, S.P. Sci. Rep. 6, 24356 (2016).
Wilkening, S. et al. Nucleic Acids Res. 41, e65 (2013).
Fimereli, D., Detours, V. & Konopka, T. Nucleic Acids Res. 41, e86 (2013).
Wu, T.D. & Nacu, S. Bioinformatics 26, 873–881 (2010).
Acknowledgements
We thank all S.P.J. laboratory members for discussions, especially A. Blackford, F. Puddu, C. Schmidt and P. Marco-Casanova for critical reading of the manuscript, and C. Le Sage and T.-W. Chiang for advice with CRISPR–Cas9 gene editing. We thank M. Leeb for H129-3 cells and advice on haploid ES cell culture conditions, and J. Hackett (Gurdon Institute, University of Cambridge) for advice in generating stable ES cell lines. We thank C.D. Robles-Espinoza for helping to design the array of baits for the exon-capture experiment, and J. Hewinson for technical support. Research in the S.P.J. laboratory is funded by Cancer Research UK (CRUK; programme grant C6/A11224), the European Research Council and the European Community Seventh Framework Programme (grant agreement no. HEALTH-F2-2010-259893; DDResponse). Core funding is provided by Cancer Research UK (C6946/A14492) and the Wellcome Trust (WT092096). S.P.J. receives salary from the University of Cambridge, supplemented by CRUK. J.V.F. was funded by Cancer Research UK programme grant C6/A11224 and the Ataxia Telangiectasia Society. J.C. was funded by Cancer Research UK programme grant C6/A11224. D.J.A. is supported by CRUK. Research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) and ERC grant agreement no. (311166). B.V.G. is supported by a Boehringer Ingelheim Fonds PhD fellowship.
Author information
Authors and Affiliations
Contributions
J.V.F. and S.P.J. designed the project. J.V.F. mutagenized haploid cells, performed 6-TG selection and isolated suppressor clones. J.V.F. and J.C. expanded suppressor clones, isolated gDNA and prepared samples for sequencing. M.H. analyzed DNA sequencing data, supervised by T.M.K. and D.J.A. J.V.F. and J.C. produced stable cell lines and CRISPR–Cas9 knock-ins. J.V.F. and J.C. isolated RNA from suppressor clones and prepared samples for sequencing. B.V.G. produced RNA sequencing libraries and T.K. analyzed RNA sequencing data, supervised by S.M.N. J.V.F. and S.P.J. wrote the manuscript, with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–7 (PDF 3132 kb)
Supplementary Data Set 1
Homozygous mutations identified through whole-exome sequencing of 7 suppressor clones. (XLSX 69 kb)
Supplementary Data Set 2
Homozygous mutations identified on the targeted exon-capture experiment performed on 189 suppressor clones. Heterozygous mutations affecting Dnmt1, Hprt, Mlh1, Msh2, Msh6 and Pms2 are also shown. (XLSX 104 kb)
Supplementary Data Set 3
Homozygous mutations identified through whole-exome sequencing of 66 suppressor clones (23 orphan clones plus 43 clones with identified mutations). Heterozygous mutations affecting Dnmt1, Hprt, Mlh1, Msh2, Msh6 and Pms2 are also shown. (XLSX 283 kb)
Supplementary Data Set 4
RNA sequencing data from 5 wild-type samples, 5 identified suppressor clones and 21 unidentified suppressor clones. Values represent fragments per kilobase per million reads. (XLSX 6230 kb)
Supplementary Data Set 5
DNA sequencing coverage for the whole-exome and targeted exon-capture experiments. (XLSX 83 kb)
Supplementary Data Set 6
R scripts for RNA sequencing analysis. (ZIP 3 kb)
Rights and permissions
About this article
Cite this article
Forment, J., Herzog, M., Coates, J. et al. Genome-wide genetic screening with chemically mutagenized haploid embryonic stem cells. Nat Chem Biol 13, 12–14 (2017). https://doi.org/10.1038/nchembio.2226
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2226
This article is cited by
-
Crimean–Congo haemorrhagic fever virus uses LDLR to bind and enter host cells
Nature Microbiology (2024)
-
Revolutionizing DNA repair research and cancer therapy with CRISPR–Cas screens
Nature Reviews Molecular Cell Biology (2023)
-
Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders
Nature Chemical Biology (2023)
-
Directed evolution in mammalian cells
Nature Methods (2021)
-
Current advances in haploid stem cells
Protein & Cell (2020)