Abstract
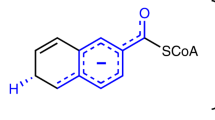
A 2.4-Å-resolution X-ray crystal structure of the carrier-protein-independent halogenase WelO5 in complex with its welwitindolinone precursor substrate, 12-epi-fischerindole U, reveals that the C13 chlorination target is proximal to the anticipated site of the oxo group in a presumptive cis-halo-oxo-iron(IV) (haloferryl) intermediate. Prior study of related halogenases forecasts substrate hydroxylation in this active-site configuration, but X-ray crystallographic verification of C13 halogenation in single crystals mandates that ligand dynamics must reposition the oxygen ligand to enable the observed outcome. S189A WelO5 produces a mixture of halogenation and hydroxylation products, showing that an outer-sphere hydrogen-bonding group orchestrates ligand movements to achieve a configuration that promotes halogen transfer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vaillancourt, F.H., Yeh, E., Vosburg, D.A., Garneau-Tsodikova, S. & Walsh, C.T. Nature's inventory of halogenation catalysts: oxidative strategies predominate. Chem. Rev. 106, 3364–3378 (2006).
Vaillancourt, F.H., Yeh, E., Vosburg, D.A., O'Connor, S.E. & Walsh, C.T. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature 436, 1191–1194 (2005).
Vaillancourt, F.H., Yin, J. & Walsh, C.T. SyrB2 in syringomycin E biosynthesis is a nonheme Fe(II) α-ketoglutarate- and O2-dependent halogenase. Proc. Natl. Acad. Sci. USA 102, 10111–10116 (2005).
Jiang, W. et al. Biosynthetic chlorination of the piperazate residue in kutzneride biosynthesis by KthP. Biochemistry 50, 6063–6072 (2011).
Galonić, D.P., Barr, E.W., Walsh, C.T., Bollinger, J.M. Jr. & Krebs, C. Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nat. Chem. Biol. 3, 113–116 (2007).
Galonić, D.P., Vaillancourt, F.H. & Walsh, C.T. Halogenation of unactivated carbon centers in natural product biosynthesis: trichlorination of leucine during barbamide biosynthesis. J. Am. Chem. Soc. 128, 3900–3901 (2006).
Neumann, C.S. & Walsh, C.T. Biosynthesis of (–)-(1S,2R)-allocoronamic acyl thioester by an Fe(II)-dependent halogenase and a cyclopropane-forming flavoprotein. J. Am. Chem. Soc. 130, 14022–14023 (2008).
Gu, L. et al. Metamorphic enzyme assembly in polyketide diversification. Nature 459, 731–735 (2009).
Pratter, S.M. et al. More than just a halogenase: modification of fatty acyl moieties by a trifunctional metal enzyme. ChemBioChem 15, 567–574 (2014).
Podgoršek, A., Zupan, M. & Iskra, J. Oxidative halogenation with “green” oxidants: oxygen and hydrogen peroxide. Angew. Chem. Int. Edn. Engl. 48, 8424–8450 (2009).
Matthews, M.L. et al. Substrate-triggered formation and remarkable stability of the C-H bond-cleaving chloroferryl intermediate in the aliphatic halogenase, SyrB2. Biochemistry 48, 4331–4343 (2009).
Bollinger, J.M. Jr. et al. Mechanisms of 2-oxoglutarate-dependent oxygenases: the hydroxylation paradigm and beyond. in 2-Oxoglutarate-Dependent Oxygenases (eds. Hausinger, R.P. & Schofield, C.J.) 95–122 (The Royal Society of Chemistry, London, 2015).
Krebs, C., Galonić Fujimori, D., Walsh, C.T. & Bollinger, J.M. Jr. Non-heme Fe(IV)-oxo intermediates. Acc. Chem. Res. 40, 484–492 (2007).
Solomon, E.I. et al. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem. Rev. 100, 235–350 (2000).
Hausinger, R.P. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 39, 21–68 (2004).
Blasiak, L.C., Vaillancourt, F.H., Walsh, C.T. & Drennan, C.L. Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature 440, 368–371 (2006).
Khare, D. et al. Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis. Proc. Natl. Acad. Sci. USA 107, 14099–14104 (2010).
Que, L. Jr. One motif—many different reactions. Nat. Struct. Biol. 7, 182–184 (2000).
Matthews, M.L. et al. Substrate positioning controls the partition between halogenation and hydroxylation in the aliphatic halogenase, SyrB2. Proc. Natl. Acad. Sci. USA 106, 17723–17728 (2009).
Wong, S.D. et al. Elucidation of the Fe(IV)=O intermediate in the catalytic cycle of the halogenase SyrB2. Nature 499, 320–323 (2013).
Martinie, R.J. et al. Experimental correlation of substrate position with reaction outcome in the aliphatic halogenase, SyrB2. J. Am. Chem. Soc. 137, 6912–6919 (2015).
Kulik, H.J. & Drennan, C.L. Substrate placement influences reactivity in non-heme Fe(II) halogenases and hydroxylases. J. Biol. Chem. 288, 11233–11241 (2013).
Borowski, T., Noack, H., Radoń, M., Zych, K. & Siegbahn, P.E. Mechanism of selective halogenation by SyrB2: a computational study. J. Am. Chem. Soc. 132, 12887–12898 (2010).
Pandian, S., Vincent, M.A., Hillier, I.H. & Burton, N.A. Why does the enzyme SyrB2 chlorinate, but does not hydroxylate, saturated hydrocarbons? A density functional theory (DFT) study. Dalton Trans. (31): 6201–6207 (2009).
Hillwig, M.L. & Liu, X. A new family of iron-dependent halogenases acts on freestanding substrates. Nat. Chem. Biol. 10, 921–923 (2014).
Hillwig, M.L. et al. Identification and characterization of a welwitindolinone alkaloid biosynthetic gene cluster in the stigonematalean Cyanobacterium Hapalosiphon welwitschii. ChemBioChem 15, 665–669 (2014).
Aik, W., McDonough, M.A., Thalhammer, A., Chowdhury, R. & Schofield, C.J. Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases. Curr. Opin. Struct. Biol. 22, 691–700 (2012).
Wong, C., Fujimori, D.G., Walsh, C.T. & Drennan, C.L. Structural analysis of an open active site conformation of nonheme iron halogenase CytC3. J. Am. Chem. Soc. 131, 4872–4879 (2009).
Hillwig, M.L., Zhu, Q. & Liu, X. Biosynthesis of ambiguine indole alkaloids in cyanobacterium Fischerella ambigua. ACS Chem. Biol. 9, 372–377 (2014).
Hillwig, M.L., Zhu, Q., Ittiamornkul, K. & Liu, X. Discovery of a promiscuous non-heme iron halogenase in ambiguine alkaloid biogenesis: implication for an evolvable enzyme family for late-stage halogenation of aliphatic carbons in small molecules. Angew. Chem. Int. Edn. Engl. 55, 5780–5784 (2016).
Zhu, Q., Hillwig, M.L., Doi, Y. & Liu, X. Aliphatic halogenase enables late-stage C-H functionalization: selective synthesis of a brominated Fischerindole alkaloid with enhanced antibacterial activity. ChemBioChem 17, 466–470 (2016).
Zhang, Z. et al. Crystal structure of a clavaminate synthase-Fe(II)-2-oxoglutarate-substrate-NO complex: evidence for metal centered rearrangements. FEBS Lett. 517, 7–12 (2002).
Chang, W.c. et al. Mechanism of the C5 stereoinversion reaction in the biosynthesis of carbapenem antibiotics. Science 343, 1140–1144 (2014).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007).
Langer, G., Cohen, S.X., Lamzin, V.S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Ten Eyck, L.F. Fast Fourier transform calculation of electron density maps. Methods Enzymol. 115, 324–337 (1985).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Schüttelkopf, A.W. & van Aalten, D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
We thank R.J. Martinie, C. Krebs, and J.M. Bollinger, Jr. for critical discussions and reading of the manuscript. We acknowledge D. Smith, J. Brunzelle, and the staff at the CCP4/APS School for Macromolecular Crystallography for assistance with X-ray data collection and analysis. This work has been supported by US National Institutes of Health grant GM100011 (A.K.B.), the Searle Scholars Program (A.K.B.), and the Department of Chemistry at the University of Pittsburgh (X.L.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). GM/CA CAT has been funded in whole or in part with US federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104).
Author information
Authors and Affiliations
Contributions
A.J.M., A.O.M., and N.R.A. purified and crystallized protein samples and collected X-ray data sets. A.J.M. and A.O.M. processed X-ray data sets and solved structures. Q.Z. and M.L.H. prepared samples and performed biochemical experiments. A.J.M., X.L., and A.K.B. designed the experimental approach, analyzed data, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.J.M., X.L., and A.K.B. have filed a patent application using information gained from this contribution.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–4 and Supplementary Figures 1–20. (PDF 12955 kb)
Rights and permissions
About this article
Cite this article
Mitchell, A., Zhu, Q., Maggiolo, A. et al. Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5. Nat Chem Biol 12, 636–640 (2016). https://doi.org/10.1038/nchembio.2112
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2112
This article is cited by
-
Algorithm-aided engineering of aliphatic halogenase WelO5* for the asymmetric late-stage functionalization of soraphens
Nature Communications (2022)
-
Reaction pathway engineering converts a radical hydroxylase into a halogenase
Nature Chemical Biology (2022)
-
Aryl C-H iodination: are there actual flavin-dependent iodinases in nature?
Science China Chemistry (2021)
-
The chloroalkaloid (−)-acutumine is biosynthesized via a Fe(II)- and 2-oxoglutarate-dependent halogenase in Menispermaceae plants
Nature Communications (2020)
-
A family of radical halogenases for the engineering of amino-acid-based products
Nature Chemical Biology (2019)