Abstract
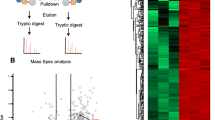
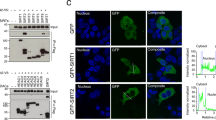
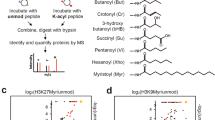
Mammalian sirtuin 6 (SIRT6) exhibits many pivotal functions and multiple enzymatic activities, but the contribution of each activity to the various functions is unclear. We identified a SIRT6 mutant (G60A) that possesses efficient defatty-acylase activity but has substantially decreased deacetylase activity in vitro and no detectable deacetylase activity in cells. The G60A mutant has a decreased ability to bind NAD+, but the presence of fatty-acyl lysine peptides restores NAD+ binding, explaining the retention of the defatty-acylase activity. Using this mutant, we found that the defatty-acylase activity of SIRT6 regulates the secretion of numerous proteins. Notably, many ribosomal proteins were secreted via exosomes from Sirt6 knockout mouse embryonic fibroblasts, and these exosomes increased NIH 3T3 cell proliferation compared with control exosomes. Our data indicate that distinct activities of SIRT6 regulate different pathways and that the G60A mutant is a useful tool to study the contribution of defatty-acylase activity to SIRT6's various functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Imai, S., Armstrong, C.M., Kaeberlein, M. & Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 (2000).
Mostoslavsky, R. et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 (2006).
Michishita, E. et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 (2008).
Mao, Z. et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446 (2011).
Kaidi, A., Weinert, B.T., Choudhary, C. & Jackson, S.P. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329, 1348–1353 (2010).
Sebastián, C. et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151, 1185–1199 (2012).
Zhong, L. et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140, 280–293 (2010).
Kawahara, T.L. et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136, 62–74 (2009).
Kanfi, Y. et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221 (2012).
Michishita, E. et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666 (2009).
Schwer, B. et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc. Natl. Acad. Sci. USA 107, 21790–21794 (2010).
Yang, B., Zwaans, B.M., Eckersdorff, M. & Lombard, D.B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8, 2662–2663 (2009).
Feldman, J.L., Baeza, J. & Denu, J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288, 31350–31356 (2013).
Jiang, H. et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 (2013).
Liszt, G., Ford, E., Kurtev, M. & Guarente, L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313–21320 (2005).
Gil, R., Barth, S., Kanfi, Y. & Cohen, H.Y. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res. 41, 8537–8545 (2013).
Du, J., Jiang, H. & Lin, H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry 48, 2878–2890 (2009).
Pan, P.W. et al. Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587 (2011).
Tennen, R.I., Berber, E. & Chua, K.F. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech. Ageing Dev. 131, 185–192 (2010).
Al-Nedawi, K., Meehan, B. & Rak, J. Microvesicles: messengers and mediators of tumor progression. Cell Cycle 8, 2014–2018 (2009).
Antonyak, M.A. et al. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. USA 108, 4852–4857 (2011).
Costa-Silva, B. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826 (2015).
Ewald, C.Y., Landis, J.N., Porter Abate, J., Murphy, C.T. & Blackwell, T.K. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519, 97–101 (2015).
Zhu, A.Y. et al. Plasmodium falciparum Sir2A preferentially hydrolyzes medium and long chain fatty acyl lysine. ACS Chem. Biol. 7, 155–159 (2012).
Charron, G. et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 131, 4967–4975 (2009).
Jiang, H., Kim, J.H., Frizzell, K.M., Kraus, W.L. & Lin, H. Clickable NAD analogues for labeling substrate proteins of poly(ADP-ribose) polymerases. J. Am. Chem. Soc. 132, 9363–9372 (2010).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Jedrusik-Bode, M. et al. The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J. Cell Sci. 126, 5166–5177 (2013).
Huang, Z. et al. Tumor suppressor Alpha B-crystallin (CRYAB) associates with the cadherin/catenin adherens junction and impairs NPC progression-associated properties. Oncogene 31, 3709–3720 (2012).
Soo, C.Y. et al. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology 136, 192–197 (2012).
Acknowledgements
We thank R. Mostoslavsky (Massachusetts General Hospital) for providing the Sirt6 WT and KO MEFs, P. Chen (Cornell University) for the use of the fluorescence spectrophotometer and S. Zhang (proteomic and MS facility of Cornell University) for help with the SILAC experiments. SILAC experiments performed on a mass spectrometer were supported by a US National Institutes of Health (NIH) SIG grant 1S10 OD017992-01. Imaging data were acquired in the Cornell BRC imaging facility using a Zeiss LSM880 confocal/multiphoton microscope funded by NYSTEM (CO29155) and NIH (S10OD018516). This work is supported by a grant from the NIH NIGMS (R01 GM098596 to H.L.).
Author information
Authors and Affiliations
Contributions
X.Z. and H.L. designed the research and wrote the manuscript; X.Z. performed biochemical and cellular studies; S.K. synthesized peptides for the in vitro deacylation study; H.J. made SIRT6 WT and TNF-α plasmids; M.A.A. performed exosome fractionation experiments; X.C. made HEK293T SIRT6 KO cells; N.A.S. and J.H.S. synthesized Alk14 and BODIPY-N3; X.Z., R.A.C. and H.L. analyzed the results; H.L. directed the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–5 and Supplementary Figures 1–18. (PDF 4223 kb)
Supplementary Data Set 1
Secretion SILAC in Sirt6 WT and KO MEFs. (XLSX 124 kb)
Supplementary Data Set 2
Proteins detected in all three SILACs. (XLSX 57 kb)
Supplementary Data Set 3
Secretion SILAC in Sirt6 KO MEFs and Sirt6 KO MEFsexpressing SIRT6 WT. (XLSX 118 kb)
Supplementary Data Set 4
Secretion SILAC in Sirt6 KO MEFs and Sirt6 KO MEFsexpressing SIRT6 G60A. (XLSX 108 kb)
Supplementary Data Set 5
Secretion SILAC in Sirt6 KO MEFs and Sirt6 KO MEFsexpressing SIRT6 H133Y. (XLSX 110 kb)
Rights and permissions
About this article
Cite this article
Zhang, X., Khan, S., Jiang, H. et al. Identifying the functional contribution of the defatty-acylase activity of SIRT6. Nat Chem Biol 12, 614–620 (2016). https://doi.org/10.1038/nchembio.2106
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2106
This article is cited by
-
Multiomics interrogation into HBV (Hepatitis B virus)-host interaction reveals novel coding potential in human genome, and identifies canonical and non-canonical proteins as host restriction factors against HBV
Cell Discovery (2021)
-
Sirt6 opposes glycochenodeoxycholate-induced apoptosis of biliary epithelial cells through the AMPK/PGC-1α pathway
Cell & Bioscience (2020)
-
NMT1 and NMT2 are lysine myristoyltransferases regulating the ARF6 GTPase cycle
Nature Communications (2020)
-
Differential modulation of SIRT6 deacetylase and deacylase activities by lysine-based small molecules
Molecular Diversity (2020)
-
Polydatin Alleviates Septic Myocardial Injury by Promoting SIRT6-Mediated Autophagy
Inflammation (2020)