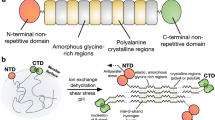
Abstract
Spider silk is strong and extensible but still biodegradable and well tolerated when implanted, making it the ultimate biomaterial. Shortcomings that arise in replicating spider silk are due to the use of recombinant spider silk proteins (spidroins) that lack native domains, the use of denaturing conditions under purification and spinning and the fact that the understanding of how spiders control silk formation is incomplete. Recent progress has unraveled the molecular mechanisms of the spidroin N- and C-terminal nonrepetitive domains (NTs and CTs) and revealed the pH and ion gradients in spiders' silk glands, clarifying how spidroin solubility is maintained and how silk is formed in a fraction of a second. Protons and CO2, generated by carbonic anhydrase, affect the stability and structures of the NT and CT in different ways. These insights should allow the design of conditions and devices for the spinning of recombinant spidroins into native-like silk.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kovoor, J. in Ecophysiology of Spiders (ed. Nentwig, W.) 160–186 (Springer-Verlag, Berlin, 1987).
Gosline, J.M., Guerette, P.A., Ortlepp, C.S. & Savage, K.N. The mechanical design of spider silks: from fibroin sequence to mechanical function. J. Exp. Biol. 202, 3295–3303 (1999).
Allmeling, C. et al. Spider silk fibres in artificial nerve constructs promote peripheral nerve regeneration. Cell Prolif. 41, 408–420 (2008).
Radtke, C. et al. Spider silk constructs enhance axonal regeneration and remyelination in long nerve defects in sheep. PLoS ONE 6, e16990 (2011).
Xia, X.X. et al. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc. Natl. Acad. Sci. USA 107, 14059–14063 (2010).This is the first report on recombinant production of native-sized spidroin.
Wong Po Foo, C. et al. Novel nanocomposites from spider silk-silica fusion (chimeric) proteins. Proc. Natl. Acad. Sci. USA 103, 9428–9433 (2006).
Teulé, F. et al. A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning. Nat. Protoc. 4, 341–355 (2009).
Rammensee, S., Slotta, U., Scheibel, T. & Bausch, A.R. Assembly mechanism of recombinant spider silk proteins. Proc. Natl. Acad. Sci. USA 105, 6590–6595 (2008).
Andersson, M. et al. Carbonic anhydrase generates CO2 and H+ that drive spider silk formation via opposite effects on the terminal domains. PLoS Biol. 12, e1001921 (2014).This paper characterizes pH regulation of spider silk formation at the molecular level.
Andersson, M., Holm, L., Ridderstrale, Y., Johansson, J. & Rising, A. Morphology and composition of the spider major ampullate gland and dragline silk. Biomacromolecules 14, 2945–2952 (2013).
Askarieh, G. et al. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 465, 236–238 (2010).This paper presents the first high-resolution structure of the NT.
Hagn, F. et al. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature 465, 239–242 (2010).This paper present the first high-resolution structure of the CT.
Kronqvist, N. et al. Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation. Nat. Commun. 5, 3254 (2014).This paper presents the molecular mechanism of NT pH-sensitive dimerization.
Ries, J., Schwarze, S., Johnson, C.M. & Neuweiler, H. Microsecond folding and domain motions of a spider silk protein structural switch. J. Am. Chem. Soc. 136, 17136–17144 (2014).
Schwarze, S., Zwettler, F.U., Johnson, C.M. & Neuweiler, H. The N-terminal domains of spider silk proteins assemble ultrafast and protected from charge screening. Nat. Commun. 4, 2815 (2013).This paper presents the molecular mechanism of ultra-fast NT association.
Gaines, W.A., Sehorn, M.G. & Marcotte, W.R. Jr. Spidroin N-terminal domain promotes a pH-dependent association of silk proteins during self-assembly. J. Biol. Chem. 285, 40745–40753 (2010).
Candelas, G.C. & Cintron, J. A spider fibron and its synthesis. J. Exp. Zool. 216, 1–6 (1981).
Rising, A. et al. Major ampullate spidroins from Euprosthenops australis: multiplicity at protein, mRNA and gene levels. Insect Mol. Biol. 16, 551–561 (2007).
Garb, J.E. & Hayashi, C.Y. Modular evolution of egg case silk genes across orb-weaving spider superfamilies. Proc. Natl. Acad. Sci. USA 102, 11379–11384 (2005).
Lane, A.K., Hayashi, C.Y., Whitworth, G.B. & Ayoub, N.A. Complex gene expression in the dragline silk producing glands of the Western black widow (Latrodectus hesperus). BMC Genomics 14, 846 (2013).
Sanggaard, K.W. et al. Spider genomes provide insight into composition and evolution of venom and silk. Nat. Commun. 5, 3765 (2014).This paper presents the first characterized spider genome.
Clarke, T.H. et al. Multi-tissue transcriptomics of the black widow spider reveals expansions, co-options, and functional processes of the silk gland gene toolkit. BMC Genomics 15, 365 (2014).
Rising, A., Hjalm, G., Engstrom, W. & Johansson, J. N-terminal nonrepetitive domain common to dragline, flagelliform, and cylindriform spider silk proteins. Biomacromolecules 7, 3120–3124 (2006).
Hayashi, C.Y., Shipley, N.H. & Lewis, R.V. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int. J. Biol. Macromol. 24, 271–275 (1999).
Vollrath, F. & Knight, D.P. Structure and function of the silk production pathway in the spider Nephila edulis. Int. J. Biol. Macromol. 24, 243–249 (1999).
Ayoub, N.A., Garb, J.E., Tinghitella, R.M., Collin, M.A. & Hayashi, C.Y. Blueprint for a high-performance biomaterial: full-length spider dragline silk genes. PLoS ONE 2, e514 (2007).This paper presents the first sequence of full-length spidroin.
Hinman, M.B. & Lewis, R.V. Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J. Biol. Chem. 267, 19320–19324 (1992).
Mita, K., Ichimura, S., Zama, M. & James, T.C. Specific codon usage pattern and its implications on the secondary structure of silk fibroin mRNA. J. Mol. Biol. 203, 917–925 (1988).
Candelas, G.C., Arroyo, G., Carrasco, C. & Dompenciel, R. Spider silkglands contain a tissue-specific alanine tRNA that accumulates in vitro in response to the stimulus for silk protein synthesis. Dev. Biol. 140, 215–220 (1990).
Chen, X., Knight, D.P. & Vollrath, F. Rheological characterization of Nephila spidroin solution. Biomacromolecules 3, 644–648 (2002).
Hijirida, D.H. et al. 13C NMR of Nephila clavipes major ampullate silk gland. Biophys. J. 71, 3442–3447 (1996).
Jin, H.J. & Kaplan, D.L. Mechanism of silk processing in insects and spiders. Nature 424, 1057–1061 (2003).
Vollrath, F. & Knight, D.P. Liquid crystalline spinning of spider silk. Nature 410, 541–548 (2001).
Hronska, M., van Beek, J.D., Williamson, P.T., Vollrath, F. & Meier, B.H. NMR characterization of native liquid spider dragline silk from Nephila edulis. Biomacromolecules 5, 834–839 (2004).
Lefèvre, T. et al. In situ conformation of spider silk proteins in the intact major ampullate gland and in solution. Biomacromolecules 8, 2342–2344 (2007).
Simmons, A.H., Michal, C.A. & Jelinski, L.W. Molecular orientation and two-component nature of the crystalline fraction of spider dragline silk. Science 271, 84–87 (1996).
van Beek, J.D., Hess, S., Vollrath, F. & Meier, B.H. The molecular structure of spider dragline silk: folding and orientation of the protein backbone. Proc. Natl. Acad. Sci. USA 99, 10266–10271 (2002).
Holland, G.P., Creager, M.S., Jenkins, J.E., Lewis, R.V. & Yarger, J.L. Determining secondary structure in spider dragline silk by carbon-carbon correlation solid-state NMR spectroscopy. J. Am. Chem. Soc. 130, 9871–9877 (2008).
Kümmerlen, J., vanBeek, J.D., Vollrath, F. & Meier, B.H. Local structure in spider dragline silk investigated by two-dimensional spin-diffusion nuclear magnetic resonance. Macromolecules 29, 2920–2928 (1996).
Holland, G.P., Jenkins, J.E., Creager, M.S., Lewis, R.V. & Yarger, J.L. Quantifying the fraction of glycine and alanine in β-sheet and helical conformations in spider dragline silk using solid-state NMR. Chem. Commun. (Camb.) 5568–5570 (2008).
Jenkins, J.E. et al. Solid-state NMR evidence for elastin-like β-turn structure in spider dragline silk. Chem. Commun. (Camb.) 46, 6714–6716 (2010).
Lindskog, S. Structure and mechanism of carbonic anhydrase. Pharmacol. Ther. 74, 1–20 (1997).
Waisbren, S.J., Geibel, J.P., Modlin, I.M. & Boron, W.F. Unusual permeability properties of gastric gland cells. Nature 368, 332–335 (1994).
Cohen, A.C. Hemolymph chemistry of two species of araneid spiders. Comp. Biochem. Physiol. 66A, 715–717 (1980).
Knight, D.P. & Vollrath, F. Changes in element composition along the spinning duct in a Nephila spider. Naturwissenschaften 88, 179–182 (2001).
Garb, J.E., Ayoub, N.A. & Hayashi, C.Y. Untangling spider silk evolution with spidroin terminal domains. BMC Evol. Biol. 10, 243 (2010).
Gao, Z. et al. Structural characterization of minor ampullate spidroin domains and their distinct roles in fibroin solubility and fiber formation. PLoS ONE 8, e56142 (2013).
Makin, O.S., Atkins, E., Sikorski, P., Johansson, J. & Serpell, L.C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. USA 102, 315–320 (2005).
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005).
Westermark, P. Aspects on human amyloid forms and their fibril polypeptides. FEBS J. 272, 5942–5949 (2005).
Jarrett, J.T. & Lansbury, P.T. Jr. Amyloid fibril formation requires a chemically discriminating nucleation event: studies of an amyloidogenic sequence from the bacterial protein OsmB. Biochemistry 31, 12345–12352 (1992).
Landreh, M. et al. A pH-dependent dimer lock in spider silk protein. J. Mol. Biol. 404, 328–336 (2010).
Jaudzems, K. et al. pH-Dependent dimerization of spider silk N-terminal domain requires relocation of a wedged tryptophan side chain. J. Mol. Biol. 422, 477–487 (2012).
Hagn, F., Thamm, C., Scheibel, T. & Kessler, H. pH-dependent dimerization and salt-dependent stabilization of the N-terminal domain of spider dragline silk—implications for fiber formation. Angew. Chem. Int. Edn Engl. 50, 310–313 (2011).
Chen, G. et al. Full-length minor ampullate spidroin gene sequence. PLoS ONE 7, e52293 (2012).
Lin, Z., Deng, Q., Liu, X.Y. & Yang, D. Engineered large spider eggcase silk protein for strong artificial fibers. Adv. Mater. 25, 1216–1220 (2013).
Stephens, J.S. et al. Effects of electrospinning and solution casting protocols on the secondary structure of a genetically engineered dragline spider silk analogue investigated via Fourier transform Raman spectroscopy. Biomacromolecules 6, 1405–1413 (2005).
Lang, G., Jokisch, S. & Scheibel, T. Air filter devices including nonwoven meshes of electrospun recombinant spider silk proteins. J. Vis. Exp. doi:10.3791/50492 (8 May 2013).
Stark, M. et al. Macroscopic fibers self-assembled from recombinant miniature spider silk proteins. Biomacromolecules 8, 1695–1701 (2007).
Xu, L., Rainey, J.K., Meng, Q. & Liu, X.Q. Recombinant minimalist spider wrapping silk proteins capable of native-like fiber formation. PLoS ONE 7, e50227 (2012).
Huemmerich, D. et al. Primary structure elements of spider dragline silks and their contribution to protein solubility. Biochemistry 43, 13604–13612 (2004).
Kallberg, Y., Gustafsson, M., Persson, B., Thyberg, J. & Johansson, J. Prediction of amyloid fibril-forming proteins. J. Biol. Chem. 276, 12945–12950 (2001).
Hessa, T. et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005).
Lefèvre, T., Paquet-Mercier, F., Rioux-Dube, J.F. & Pezolet, M. Review structure of silk by Raman spectromicroscopy: from the spinning glands to the fibers. Biopolymers 97, 322–336 (2012).
Wang, S., Huang, W. & Yang, D. NMR structure note: repetitive domain of aciniform spidroin 1 from Nephila antipodiana. J. Biomol. NMR 54, 415–420 (2012).
Blackledge, T.A. & Hayashi, C.Y. Silken toolkits: biomechanics of silk fibers spun by the orb web spider Argiope argentata (Fabricius 1775). J. Exp. Biol. 209, 2452–2461 (2006).
Denny, M. The physical properties of spider's silk and their role in the design of orb-webs. J. Exp. Biol. 65, 483–506 (1976).
Köhler, T. & Vollrath, F. Thread biomechanics in the two orb weaving spiders Araneus diadematus (Araneae, Araneidae) and Uloborus walckenaerius (Araneae, Uloboridae). J. Exp. Zool. 271, 1–17 (1995).
Stauffer, S.L., Coguill, S.L. & Lewis, R.V. Comparison of physical properties of 3 silks from Nephila clavipes and Araneus gemmoides. J. Arachnol. 22, 5–11 (1994).
Acknowledgements
We are grateful to S. Knight for valuable discussions and helpful comments on this manuscript. The Swedish Research Council supported work in the authors' laboratory.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Summary of techniques and proteins used to make artificial spider silk fibers (PDF 200 kb)
Rights and permissions
About this article
Cite this article
Rising, A., Johansson, J. Toward spinning artificial spider silk. Nat Chem Biol 11, 309–315 (2015). https://doi.org/10.1038/nchembio.1789
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1789
This article is cited by
-
Replicating shear-mediated self-assembly of spider silk through microfluidics
Nature Communications (2024)
-
Engineered spidroin-derived high-performance fibers for diverse applications
Nano Research (2024)
-
A molecular atlas reveals the tri-sectional spinning mechanism of spider dragline silk
Nature Communications (2023)
-
Aqueous spinning of robust, self-healable, and crack-resistant hydrogel microfibers enabled by hydrogen bond nanoconfinement
Nature Communications (2023)
-
Assembly of Nanowires into Macroscopic One-Dimensional Fibers in Liquid State
Advanced Fiber Materials (2023)