Abstract
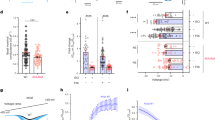
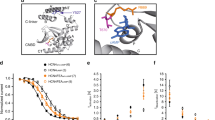
cAMP mediates autonomic regulation of heart rate by means of hyperpolarization-activated cyclic nucleotide-gated (HCN) channels, which underlie the pacemaker current If. cAMP binding to the C-terminal cyclic nucleotide binding domain enhances HCN open probability through a conformational change that reaches the pore via the C-linker. Using structural and functional analysis, we identified a binding pocket in the C-linker of HCN4. Cyclic dinucleotides, an emerging class of second messengers in mammals, bind the C-linker pocket (CLP) and antagonize cAMP regulation of the channel. Accordingly, cyclic dinucleotides prevent cAMP regulation of If in sinoatrial node myocytes, reducing heart rate by 30%. Occupancy of the CLP hence constitutes an efficient mechanism to hinder β-adrenergic stimulation on If. Our results highlight the regulative role of the C-linker and identify a potential drug target in HCN4. Furthermore, these data extend the signaling scope of cyclic dinucleotides in mammals beyond their first reported role in innate immune system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
16 June 2014
In the version of this article initially published, a data point and one of its two error bars were missing from Figure 3e in the row depicting mutant F564T. The error has been corrected in the HTML and PDF versions of the article.
References
DiFrancesco, D. Pacemaker mechanisms in cardiac tissue. Annu. Rev. Physiol. 55, 455–472 (1993).
Ludwig, A., Zong, X., Jeglitsch, M., Hofmann, F. & Biel, M. A family of hyperpolarization-activated mammalian cation channels. Nature 393, 587–591 (1998).
Santoro, B. et al. Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell 93, 717–729 (1998).
Zagotta, W.N. et al. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425, 200–205 (2003).
Craven, K.B., Olivier, N.B. & Zagotta, W.N. C-terminal movement during gating in cyclic nucleotide-modulated channels. J. Biol. Chem. 283, 14728–14738 (2008).
Clayton, G.M., Silverman, W.R., Heginbotham, L. & Morais-Cabral, J.H. Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel. Cell 119, 615–627 (2004).
Schunke, S., Stoldt, M., Novak, K., Kaupp, U.B. & Willbold, D. Solution structure of the Mesorhizobium loti K1 channel cyclic nucleotide-binding domain in complex with cAMP. EMBO Rep. 10, 729–735 (2009).
Romling, U., Galperin, M.Y. & Gomelsky, M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 (2013).
Chen, Z.H. & Schaap, P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature 488, 680–683 (2012).
Wu, J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013).
Burdette, D.L. et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011).
DiFrancesco, D. & Tortora, P. Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature 351, 145–147 (1991).
Chandler, N.J. et al. Molecular architecture of the human sinus node: insights into the function of the cardiac pacemaker. Circulation 119, 1562–1575 (2009).
Lolicato, M. et al. Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels. J. Biol. Chem. 286, 44811–44820 (2011).
Davies, B.W., Bogard, R.W., Young, T.S. & Mekalanos, J.J. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370 (2012).
Gao, P. et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013).
Diner, E.J. et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 3, 1355–1361 (2013).
Ablasser, A. et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013).
Zhang, X. et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 (2013).
Yin, Q. et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46, 735–745 (2012).
DiFrancesco, D., Ducouret, P. & Robinson, R.B. Muscarinic modulation of cardiac rate at low acetylcholine concentrations. Science 243, 669–671 (1989).
Santoro, B. et al. TRIP8b splice variants form a family of auxiliary subunits that regulate gating and trafficking of HCN channels in the brain. Neuron 62, 802–813 (2009).
Zolles, G. et al. Association with the auxiliary subunit PEX5R/Trip8b controls responsiveness of HCN channels to cAMP and adrenergic stimulation. Neuron 62, 814–825 (2009).
DiFrancesco, J.C. et al. Recessive loss-of-function mutation in the pacemaker HCN2 channel causing increased neuronal excitability in a patient with idiopathic generalized epilepsy. J. Neurosci. 31, 17327–17337 (2011).
Bucchi, A., Tognati, A., Milanesi, R., Baruscotti, M. & DiFrancesco, D. Properties of ivabradine-induced block of HCN1 and HCN4 pacemaker channels. J. Physiol. (Lond.) 572, 335–346 (2006).
Li, X.D. et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 (2013).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Uhlen, M. et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 (2010).
Zorn-Pauly, K. et al. Endotoxin impairs the human pacemaker current If . Shock 28, 655–661 (2007).
Werdan, K. et al. Impaired regulation of cardiac function in sepsis, SIRS, and MODS. Can. J. Physiol. Pharmacol. 87, 266–274 (2009).
Klöckner, U. et al. Differential reduction of HCN channel activity by various types of lipopolysaccharide. J. Mol. Cell. Cardiol. 51, 226–235 (2011).
Ablasser, A. et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534 (2013).
Leslie, A.G.M. MOSFLM User Guide, Mosflm Version 6.2.3 (MRC laboratory of Molecular Biology, Cambridge, UK, 2003).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Laskowski, R.A., Macarthur, M.W., Moss, D.S. & Thornton, J.M. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Krissinel, E. & Henrik, K. in Computational Life Sciences, Vol. 3695 (eds. Berthold, M.R., Glen, R.C., Diederichs, K., Kohlbacher, O. & Fischer, I.) 163–174 (2005).
Accili, E.A. & DiFrancesco, D. Inhibition of the hyperpolarization-activated current (If) of rabbit SA node myocytes by niflumic acid. Pflugers Arch. 431, 757–762 (1996).
DiFrancesco, D. & Mangoni, M. Modulation of single hyperpolarization-activated channels (If) by cAMP in the rabbit sino-atrial node. J. Physiol. (Lond.) 474, 473–482 (1994).
Baruscotti, M. et al. Deep bradycardia and heart block caused by inducible cardiac-specific knockout of the pacemaker channel gene Hcn4. Proc. Natl. Acad. Sci. USA 108, 1705–1710 (2011).
Bucchi, A., Baruscotti, M., Robinson, R.B. & DiFrancesco, D.I. If-dependent modulation of pacemaker rate mediated by cAMP in the presence of ryanodine in rabbit sino-atrial node cells. J. Mol. Cell. Cardiol. 35, 905–913 (2003).
Kranzusch, P.J., Lee, A.S., Berger, J.M. & Doudna, J. A. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 3, 1362–1368 (2013).
Zsoldos, Z., Szabo, I., Szabo, Z. & Johnson, A.P. Software tools for structure based rational drug design. J. Mol. Struct. THEOCHEM 666, 659–665 (2003).
Zsoldos, Z., Reid, D., Simon, A., Sadjad, B.S. & Johnson, A.P. eHiTS: an innovative approach to the docking and scoring function problems. Curr. Protein Pept. Sci. 7, 421–435 (2006).
Zsoldos, Z., Reid, D., Simon, A., Sadjad, S.B. & Johnson, A.P. eHiTS: a new fast, exhaustive flexible ligand docking system. J. Mol. Graph. Model. 26, 198–212 (2007).
Acknowledgements
We thank Xention Ltd. (Cambridge, UK) for the generous gift of HCNs cDNA, J. Berger and J.A. Doudna for hSTING cDNA and C. Deutscher (BioLog, Bremen) for technical help. This work was supported by SAL-49 Progetto di Cooperazione Scientifica e Tecnologica Regione Lombardia, Programmi di Ricerca di Rilevante Interesse Nazionale 2010CSJX4F and Ministero Affari Esteri 01467532013-06-27 to A.M.; by European Drug Initiative on Channels and Transporters to K.S., A.P.J., C.W.G.F. and A.M; and by Bundesministerium für Bildung und Forschung (GREVIS) project to G.T.
Author information
Authors and Affiliations
Contributions
M.L. purified, crystallized, collected, processed and refined X-ray data. M.N. processed and refined X-ray data, prepared some of the figures and, with M.B., revised the manuscript. C.A. prepared the mutants and, with M.A., performed the whole-cell experiments. S.Z. and I.S. performed the inside-out experiments. A.B. made measurements and, with D.D., designed and analyzed the experiments on mouse SAN cells. K.S. performed the docking studies with guidance from A.P.J. and C.W.G.F. F.S. and D.K. synthesized cyclic dinucleotides and contributed information on their signaling properties. G.T. and A.M. designed the study, analyzed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
F.S. and D.K. are Head of Research and Development and staff member, respectively, at the BIOLOG Life Science Institute, which sells cyclic nucleotide analogs for research purposes.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–8 and Supplementary Tables 1–3. (PDF 1265 kb)
Rights and permissions
About this article
Cite this article
Lolicato, M., Bucchi, A., Arrigoni, C. et al. Cyclic dinucleotides bind the C-linker of HCN4 to control channel cAMP responsiveness. Nat Chem Biol 10, 457–462 (2014). https://doi.org/10.1038/nchembio.1521
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1521
This article is cited by
-
Activation of CB1R alleviates central sensitization by regulating HCN2-pNR2B signaling in a chronic migraine rat model
The Journal of Headache and Pain (2023)
-
Discrimination between cyclic nucleotides in a cyclic nucleotide-gated ion channel
Nature Structural & Molecular Biology (2023)
-
Sinoatrial node pacemaker cells share dominant biological properties with glutamatergic neurons
Protein & Cell (2021)
-
A mutant HCN4 channel in a family with bradycardia, left bundle branch block, and left ventricular noncompaction
Heart and Vessels (2018)
-
Versatile modes of cellular regulation via cyclic dinucleotides
Nature Chemical Biology (2017)