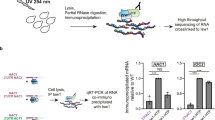
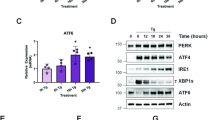
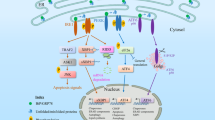
Abstract
Accumulation of unfolded proteins within the endoplasmic reticulum (ER) of eukaryotic cells leads to an unfolded protein response (UPR) that either restores homeostasis or commits the cells to apoptosis. Tools traditionally used to study the UPR are proapoptotic and thus confound analysis of long-term cellular responses to ER stress. Here, we describe an ER-localized HaloTag (ERHT) protein that can be conditionally destabilized using a small-molecule hydrophobic tag (HyT36). Treatment of ERHT-expressing cells with HyT36 induces acute, resolvable ER stress that results in transient UPR activation without induction of apoptosis. Transcriptome analysis of late-stage responses to this UPR stimulus reveals a link between UPR activity and estrogen signaling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Shoulders, M.D. et al. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 3, 1279–1292 (2013).
Smith, M.H., Ploegh, H.L. & Weissman, J.S. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334, 1086–1090 (2011).
Bernasconi, R., Galli, C., Kokame, K. & Molinari, M. Autoadaptive ER-associated degradation defines a preemptive unfolded protein response pathway. Mol. Cell 52, 783–793 (2013).
Mori, K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J. Biochem. 146, 743–750 (2009).
Tirasophon, W., Welihinda, A.A. & Kaufman, R.J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12, 1812–1824 (1998).
Korennykh, A.V. et al. The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693 (2009).
Calfon, M. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002).
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011).
Coelho, D.S. et al. Xbp1-independent Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in Drosophila. Cell Rep. 5, 791–801 (2013).
Han, D. et al. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138, 562–575 (2009).
Tabas, I. & Ron, D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190 (2011).
Haze, K., Yoshida, H., Yanagi, H., Yura, T. & Mori, K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 (1999).
Bertolotti, A., Zhang, Y., Hendershot, L.M., Harding, H.P. & Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326–332 (2000).
Liu, C.Y., Schroder, M. & Kaufman, R.J. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 275, 24881–24885 (2000).
Harding, H.P. et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 (2000).
Harding, H.P., Zhang, Y., Bertolotti, A., Zeng, H. & Ron, D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 (2000).
Han, J. et al. ER-stress–induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 (2013).
Hetz, C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102 (2012).
Lytton, J., Westlin, M. & Hanley, M.R. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J. Biol. Chem. 266, 17067–17071 (1991).
Takatsuki, A., Arima, K. & Tamura, G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J. Antibiot. (Tokyo) 24, 215–223 (1971).
Helms, J.B. & Rothman, J.E. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature 360, 352–354 (1992).
Carlberg, M. et al. Short exposures to tunicamycin induce apoptosis in SV40-transformed but not in normal human fibroblasts. Carcinogenesis 17, 2589–2596 (1996).
Kaufman, R.J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211–1233 (1999).
Lawless, M.W. et al. Activation of endoplasmic reticulum-specific stress responses associated with the conformational disease Z α1-antitrypsin deficiency. J. Immunol. 172, 5722–5726 (2004).
Hidvegi, T., Schmidt, B.Z., Hale, P. & Perlmutter, D.H. Accumulation of mutant α1-antitrypsin Z in the endoplasmic reticulum activates caspases-4 and -12, NFκB, and BAP31 but not the unfolded protein response. J. Biol. Chem. 280, 39002–39015 (2005).
Mori, A. et al. Derlin-1 overexpression ameliorates mutant SOD1-induced endoplasmic reticulum stress by reducing mutant SOD1 accumulation. Neurochem. Int. 58, 344–353 (2011).
Nadanaka, S., Yoshida, H., Kano, F., Murata, M. & Mori, K. Activation of mammalian unfolded protein response is compatible with the quality control system operating in the endoplasmic reticulum. Mol. Biol. Cell 15, 2537–2548 (2004).
Sellmyer, M.A., Chen, L.C., Egeler, E.L., Rakhit, R. & Wandless, T.J. Intracellular context affects levels of a chemically dependent destabilizing domain. PLoS ONE 7, e43297 (2012).
Neklesa, T.K. et al. A bidirectional system for the dynamic small molecule control of intracellular fusion proteins. ACS Chem. Biol. 8, 2293–2300 (2013).
Tae, H.S. et al. Identification of hydrophobic tags for the degradation of stabilized proteins. ChemBioChem 13, 538–541 (2012).
Neklesa, T.K. et al. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat. Chem. Biol. 7, 538–543 (2011).
Munro, S. & Pelham, H.R. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 (1987).
Pelham, H.R. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 7, 913–918 (1988).
Manders, E. Measurement of colocalization of objects in dual-color confocal images. J. Microsc. 169, 375–382 (1993).
Gardner, B.M. & Walter, P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333, 1891–1894 (2011).
Wang, Y. et al. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275, 27013–27020 (2000).
Lu, M., Mira-y-Lopez, R., Nakajo, S., Nakaya, K. & Jing, Y. Expression of estrogen receptor α, retinoic acid receptor α and cellular retinoic acid binding protein II genes is coordinately regulated in human breast cancer cells. Oncogene 24, 4362–4369 (2005).
Li, Q. et al. Identification and implantation stage-specific expression of an interferon-α–regulated gene in human and rat endometrium. Endocrinology 142, 2390–2400 (2001).
Thompson, C.J., Tam, N.N., Joyce, J.M., Leav, I. & Ho, S.M. Gene expression profiling of testosterone and estradiol-17 β-induced prostatic dysplasia in Noble rats and response to the antiestrogen ICI 182,780. Endocrinology 143, 2093–2105 (2002).
Mercier, I. et al. Genetic ablation of caveolin-1 drives estrogen-hypersensitivity and the development of DCIS-like mammary lesions. Am. J. Pathol. 174, 1172–1190 (2009).
Han, X. et al. Role of estrogen receptor α and β in preserving hippocampal function during aging. J. Neurosci. 33, 2671–2683 (2013).
Pedram, A., Razandi, M., Aitkenhead, M., Hughes, C.C. & Levin, E.R. Integration of the non-genomic and genomic actions of estrogen. Membrane-initiated signaling by steroid to transcription and cell biology. J. Biol. Chem. 277, 50768–50775 (2002).
Levin, E.R. Bidirectional signaling between the estrogen receptor and the epidermal growth factor receptor. Mol. Endocrinol. 17, 309–317 (2003).
Ding, L. et al. Ligand-independent activation of estrogen receptor α by XBP-1. Nucleic Acids Res. 31, 5266–5274 (2003).
Papa, L. & Germain, D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J. Cell Sci. 124, 1396–1402 (2011).
Haynes, C.M. & Ron, D. The mitochondrial UPR—protecting organelle protein homeostasis. J. Cell Sci. 123, 3849–3855 (2010).
Siegelin, M.D. et al. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J. Clin. Invest. 121, 1349–1360 (2011).
Kang, B.H. et al. Preclinical characterization of mitochondria-targeted small molecule hsp90 inhibitors, gamitrinibs, in advanced prostate cancer. Clin. Cancer Res. 16, 4779–4788 (2010).
Sin, N. et al. Total synthesis of the potent proteasome inhibitor epoxomicin: a useful tool for understanding proteasome biology. Bioorg. Med. Chem. Lett. 9, 2283–2288 (1999).
Krämer, A., Green, J., Pollard, J. Jr. & Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014).
Acknowledgements
We wish to acknowledge financial support from the US National Institutes of Health (R01AI084140, R01CA083049 and T32GM067543) and the Department of Defense through the National Defense Science and Engineering Graduate Fellowship Program.
Author information
Authors and Affiliations
Contributions
K.R. and D.J.N. contributed equally as first authors to this work. C.M.C. conceived the idea. K.R. generated the ERHT construct and cell line and performed the immunoprecipitation and immunoblotting studies. D.J.N., K.R., Y.V.S. and A.A. performed the qPCR experiments. D.J.N. analyzed the RNA-seq data. Y.V.S. and D.J.N. assessed compound cytotoxicity. C.Z. and Y.V.S. performed the ERAD experiments. D.J.N., K.R. and C.M.C. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–23 and Supplementary Tables 1–3. (PDF 2195 kb)
Supplementary Data Set 1
Mean fold upregulation of genes from two independent mRNA-sequencing experiments: full gene list used for analysis using Ingenuity Pathway Analysis (IPA) is provided. (XLSX 1181 kb)
Rights and permissions
About this article
Cite this article
Raina, K., Noblin, D., Serebrenik, Y. et al. Targeted protein destabilization reveals an estrogen-mediated ER stress response. Nat Chem Biol 10, 957–962 (2014). https://doi.org/10.1038/nchembio.1638
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1638
This article is cited by
-
Downregulation of EphA2 stability by RNF5 limits its tumor-suppressive function in HER2-negative breast cancers
Cell Death & Disease (2023)
-
Hair androgen concentrations and depressive disorders in adolescents from the general population
European Child & Adolescent Psychiatry (2023)
-
Hypoxia-induced SETX links replication stress with the unfolded protein response
Nature Communications (2021)
-
Targeted protein degradation as a powerful research tool in basic biology and drug target discovery
Nature Structural & Molecular Biology (2020)
-
Quantification of very low-abundant proteins in bacteria using the HaloTag and epi-fluorescence microscopy
Scientific Reports (2019)