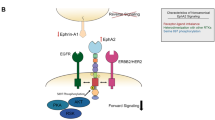
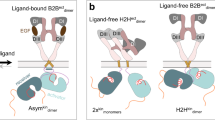
Abstract
Aberrant signaling through the Axl receptor tyrosine kinase has been associated with a myriad of human diseases, most notably metastatic cancer, identifying Axl and its ligand Gas6 as important therapeutic targets. Using rational and combinatorial approaches, we engineered an Axl 'decoy receptor' that binds Gas6 with high affinity and inhibits its function, offering an alternative approach from drug discovery efforts that directly target Axl. Four mutations within this high-affinity Axl variant caused structural alterations in side chains across the Gas6-Axl binding interface, stabilizing a conformational change on Gas6. When reformatted as an Fc fusion, the engineered decoy receptor bound Gas6 with femtomolar affinity, an 80-fold improvement compared to binding of the wild-type Axl receptor, allowing effective sequestration of Gas6 and specific abrogation of Axl signaling. Moreover, increased Gas6 binding affinity was critical and correlative with the ability of decoy receptors to potently inhibit metastasis and disease progression in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lemke, G. & Rothlin, C.V. Immunobiology of the TAM receptors. Nat. Rev. Immunol. 8, 327–336 (2008).
Li, Y. et al. Axl as a potential therapeutic target in cancer: role of Axl in tumor growth, metastasis and angiogenesis. Oncogene 28, 3442–3455 (2009).
Zhang, Z. et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 44, 852–860 (2012).
Hong, J., Peng, D., Chen, Z., Sehdev, V. & Belkhiri, A. ABL regulation by AXL promotes cisplatin resistance in esophageal cancer. Cancer Res. 73, 331–340 (2013).
Byers, L.A. et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 19, 279–290 (2013).
Vajkoczy, P. et al. Dominant-negative inhibition of the Axl receptor tyrosine kinase suppresses brain tumor cell growth and invasion and prolongs survival. Proc. Natl. Acad. Sci. USA 103, 5799–5804 (2006).
Linger, R.M., Keating, A.K., Earp, H.S. & Graham, D.K. TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv. Cancer Res. 100, 35–83 (2008).
Gustafsson, A. et al. Differential expression of Axl and Gas6 in renal cell carcinoma reflecting tumor advancement and survival. Clin. Cancer Res. 15, 4742–4749 (2009).
Chen, P.X., Li, Q.Y. & Yang, Z. Axl and prostasin are biomarkers for prognosis of ovarian adenocarcinoma. Ann. Diagn. Pathol. 17, 425–429 (2013).
Rankin, E.B. et al. AXL is an essential factor and therapeutic target for metastatic ovarian cancer. Cancer Res. 70, 7570–7579 (2010).
Holland, S.J. et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 70, 1544–1554 (2010).
Yan, S.B. et al. LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models. Invest. New Drugs 31, 833–844 (2013).
Ye, X. et al. An anti-Axl monoclonal antibody attenuates xenograft tumor growth and enhances the effect of multiple anticancer therapies. Oncogene 29, 5254–5264 (2010).
Leconet, W. et al. Preclinical validation of AXL receptor as a target for antibody-based pancreatic cancer immunotherapy. Oncogene 10.1038/onc.2013.487 (2013).
Cerchia, L. et al. Targeting Axl with an high-affinity inhibitory aptamer. Mol. Ther. 20, 2291–2303 (2012).
Paolino, M. et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 507, 508–512 (2014).
Fabian, M.A. et al. A small molecule–kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 23, 329–336 (2005).
Fisher, P.W. et al. A novel site contributing to growth-arrest–specific gene 6 binding to its receptors as revealed by a human monoclonal antibody. Biochem. J. 387, 727–735 (2005).
Varnum, B.C. et al. Axl receptor tyrosine kinase stimulated by the vitamin-K–dependent protein encoded by growth-arrest-specific gene-6. Nature 373, 623–626 (1995).
Meyer, A.S., Miller, M.A., Gertler, F.B. & Lauffenburger, D.A. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci. Signal. 6, ra66 (2013).
Kariolis, M.S., Kapur, S. & Cochran, J.R. Beyond antibodies: using biological principles to guide the development of next-generation protein therapeutics. Curr. Opin. Biotechnol. 24, 1072–1077 (2013).
Economides, A.N. et al. Cytokine traps: multi-component, high-affinity blockers of cytokine action. Nat. Med. 9, 47–52 (2003).
Peppel, K., Crawford, D. & Beutler, B. A tumor necrosis factor (TNF) receptor-IgG heavy chain chimeric protein as a bivalent antagonist of TNF activity. J. Exp. Med. 174, 1483–1489 (1991).
Holash, J. et al. VEGF-Trap: a VEGF blocker with potent antitumor effects. Proc. Natl. Acad. Sci. USA 99, 11393–11398 (2002).
Sasaki, T. et al. Structural basis for Gas6-Axl signalling. EMBO J. 25, 80–87 (2006).
Darling, R.J. & Brault, P.A. Kinetic exclusion assay technology: characterization of molecular interactions. Assay Drug Dev. Technol. 2, 647–657 (2004).
Hafizi, S. & Dahlback, B. Gas6 and protein S. Vitamin K–dependent ligands for the Axl receptor tyrosine kinase subfamily. FEBS J. 273, 5231–5244 (2006).
Manfioletti, G., Brancolini, C., Avanzi, G. & Schneider, C. The protein encoded by a growth arrest-specific gene (gas6) is a new member of the vitamin K–dependent proteins related to protein S, a negative coregulator in the blood coagulation cascade. Mol. Cell. Biol. 13, 4976–4985 (1993).
Sasaki, T. et al. Crystal structure of a C-terminal fragment of growth arrest-specific protein Gas6—receptor tyrosine kinase activation by laminin G–like domains. J. Biol. Chem. 277, 44164–44170 (2002).
Chakrabartty, A., Doig, A.J. & Baldwin, R.L. Helix capping propensities in peptides parallel those in proteins. Proc. Natl. Acad. Sci. USA 90, 11332–11336 (1993).
Levin, A.M. et al. Exploiting a natural conformational switch to engineer an interleukin-2 'superkine'. Nature 484, 529–533 (2012).
Marshall, S.A., Lazar, G.A., Chirino, A.J. & Desjarlais, J.R. Rational design and engineering of therapeutic proteins. Drug Discov. Today 8, 212–221 (2003).
Gjerdrum, C. et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc. Natl. Acad. Sci. USA 107, 1124–1129 (2010).
Aslakson, C.J. & Miller, F.R. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 52, 1399–1405 (1992).
Wan, L., Pantel, K. & Kang, Y. Tumor metastasis: moving new biological insights into the clinic. Nat. Med. 19, 1450–1464 (2013).
Lu, Q. et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398, 723–728 (1999).
Angelillo-Scherrer, A. et al. Deficiency or inhibition of Gas6 causes platelet dysfunction and protects mice against thrombosis. Nat. Med. 7, 215–221 (2001).
Paolino, M. et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 507, 508–512 (2014).
Boder, E.T. & Wittrup, K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997).
Chao, G. et al. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1, 755–768 (2006).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
The PyMOL Molecular Graphics System. Version 1.3r1, Schrödinger, LLC (2010).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Yuan, Z., Zhao, J. & Wang, Z.X. Flexibility analysis of enzyme active sites by crystallographic temperature factors. Protein Eng. 16, 109–114 (2003).
Acknowledgements
The authors thank E. Hohenester (Imperial College London) for his generous gift of the Gas6 plasmid and helpful discussions regarding protein production, L.L. Zheng for technical assistance with protein production and purification and the Stanford FACS Core Facility for assistance with the flow cytometric sorting. All in vivo and ex vivo imaging was conducted in the Stanford Small Animal Imaging Facility. Portions of this research were performed at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy Office of Basic Energy Sciences. This work was supported by the Wallace H. Coulter Translational Research Grant Program (J.R.C. and A.J.G.), the Stanford Chemistry, Engineering & Medicine for Human Health (ChEM-H) Institute (J.R.C. and I.I.M.), Stanford Bio-X and ARCS Graduate Fellowships (M.S.K.), US National Institutes of Health (NIH) grants CA-088480 and CA-67166 (A.J.G. and Y.R.M.), NIH National Institute of General Medical Sciences Training Grant T32 GM008412 and the Siebel Graduate Fellowship (D.S.J.).
Author information
Authors and Affiliations
Contributions
M.S.K., Y.R.M., D.S.J., S.K., I.I.M., A.J.G. and J.R.C. designed the research; M.S.K. and D.S.J. performed protein engineering and characterization; M.S.K., S.K. and I.I.M. did the crystallography; M.S.K. and Y.R.M. conducted the in vivo experiments; all of the authors analyzed the data; and M.S.K. prepared the manuscript with input from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
Stanford University holds patent number US8618254 B2, 'Biologic inhibitors for therapeutic targeting the receptor tyrosine kinase AXL', which is related to the work described in this paper, with M.S.K., Y.R.M., D.S.J., A.J.G. and J.R.C. named as inventors. A.J.G. is a founder of Ruga, a company that discovers and develops targeted therapeutics in oncology that has licensed this patent.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–14 and Supplementary Tables 1–7. (PDF 6640 kb)
Rights and permissions
About this article
Cite this article
Kariolis, M., Miao, Y., Jones, D. et al. An engineered Axl 'decoy receptor' effectively silences the Gas6-Axl signaling axis. Nat Chem Biol 10, 977–983 (2014). https://doi.org/10.1038/nchembio.1636
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1636
This article is cited by
-
Therapeutic targeting of the functionally elusive TAM receptor family
Nature Reviews Drug Discovery (2023)
-
Gas6 in chronic liver disease—a novel blood-based biomarker for liver fibrosis
Cell Death Discovery (2023)
-
TAM family kinases as therapeutic targets at the interface of cancer and immunity
Nature Reviews Clinical Oncology (2023)
-
An engineered ligand trap inhibits leukemia inhibitory factor as pancreatic cancer treatment strategy
Communications Biology (2021)
-
Regulation of efferocytosis as a novel cancer therapy
Cell Communication and Signaling (2020)