Abstract
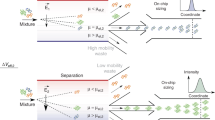
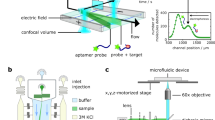
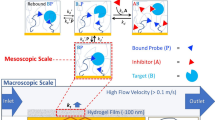
The study of biomolecular interactions is central to an understanding of function, malfunction and therapeutic modulation of biological systems, yet often involves a compromise between sensitivity and accuracy. Many conventional analytical steps and the procedures required to facilitate sensitive detection, such as the incorporation of chemical labels, are prone to perturb the complexes under observation. Here we present a ‘latent’ analysis approach that uses chemical and microfluidic tools to reveal, through highly sensitive detection of a labelled system, the behaviour of the physiologically relevant unlabelled system. We implement this strategy in a native microfluidic diffusional sizing platform, allowing us to achieve detection sensitivity at the attomole level, determine the hydrodynamic radii of biomolecules that vary by over three orders of magnitude in molecular weight, and study heterogeneous mixtures. We illustrate these key advantages by characterizing a complex of an antibody domain in the solution phase and under physiologically relevant conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008).
Suh, E. H. et al. Stilbene vinyl sulfonamides as fluorogenic sensors of and traceless covalent kinetic stabilizers of transthyretin that prevent amyloidogenesis. J. Am. Chem. Soc. 135, 17869–17880 (2013).
Aguzzi, A. & Calella, A. M. Prions: protein aggregation and infectious diseases. Phys. Rev. 89, 1105–1152 (2009).
Campioni, S. et al. The presence of an air–water interface affects formation and elongation of α-synuclein fibrils. J. Am. Chem. Soc. 136, 2866–2875 (2014).
Adamcik, J. et al. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nature Nanotech. 5, 423–428 (2010).
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Arkin, M. R. & Whitty, A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein–protein interactions. Curr. Opin. Chem. Biol. 13, 284–290 (2009).
Wells, J. A. & McClendon, C. L. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature 450, 1001–1009 (2007).
Berggård, T., Linse, S. & James, P. Methods for the detection and analysis of protein–protein interactions. Proteomics 7, 2833–2842 (2007).
Uetz, P. et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 (2000).
Ho, Y. et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 (2002).
Yu, X., Xu, D. & Cheng, Q. Label-free detection methods for protein microarrays. Proteomics 6, 5493–5503 (2006).
Zuiderweg, E. R. P. Mapping protein–protein interactions in solution by NMR spectroscopy. Biochemistry 41, 1–7 (2001).
Hanlon, A. D., Larkin, M. I. & Reddick, R. M. Free-solution, label-free protein–protein interactions characterized by dynamic light scattering. Biophys. J. 98, 297–304 (2010).
Lee, T. T. & Yeung, E. S. High-sensitivity laser-induced fluorescence detection of native proteins in capillary electrophoresis. J. Chromatogr. A 595, 319–325 (1992).
Bornhop, D. J. et al. Free-solution, label-free molecular interactions studied by back-scattering interferometry. Science 317, 1732–1736 (2007).
Jungbauer, L. M., Yu, C., Laxton, K. J. & LaDu, M. J. Preparation of fluorescently-labeled amyloid-beta peptide assemblies: the effect of fluorophore conjugation on structure and function. J. Mol. Recogn. 22, 403–413 (2009).
Herling, T. W. et al. Integration and characterization of solid wall electrodes in microfluidic devices fabricated in a single photolithography step. Appl. Phys. Lett. 102, 184102–4 (2013).
Bruus, H. Theoretical Microfluidics (Oxford Master Series in Physics, Oxford Univ. Press, 2008).
Brody, J. P. & Yager, P. Diffusion-based extraction in a microfabricated device. Sens. Actuat. A 58, 13–18 (1997).
Weigl, B. H. & Yager, P. Microfluidic diffusion-based separation and detection. Science 283, 346–347 (1999).
Kamholz, A. E., Weigl, B. H., Finlayson, B. A. & Yager, P. Quantitative analysis of molecular interaction in a microfluidic channel: the T-sensor. Anal. Chem. 71, 5340–5347 (1999).
Hatch, A. et al. A rapid diffusion immunoassay in a T-sensor. Nature Biotechol. 19, 461–465 (2001).
Kamholz, A. E., Schilling, E. A. & Yager, P. Optical measurement of transverse molecular diffusion in a microchannel. Biophys. J. 80, 1967–1972 (2001).
Knowles, T., Devlin, G., Dobson, C. & Welland, M. in Methods in Molecular Biology Vol. 752 (eds Hill, A. F., Barnham, K. J., Bottomley, S. P. & Cappai, R.) 137–145 (Humana, 2011).
Gao, D., Liu, H., Jiang, Y. & Lin, J.-M. Recent advances in microfluidics combined with mass spectrometry: technologies and applications. Lab Chip 13, 3309–3322 (2013).
Roth, M. Fluorescence reaction for amino acids. Anal. Chem. 43, 880–882 (1971).
Benson, J. R. & Hare, P. o-Phthalaldehyde fluorogenic detection of primary amines in the picomole range. comparison with fluorescamine and ninhydrin. Proc. Natl Acad. Sci. USA 72, 619–622 (1975).
Simons, S. S. & Johnson, D. F. The structure of the fluorescent adduct formed in the reaction of o-phthalaldehyde and thiols with amines. J, Am. Chem. Soc. 98, 7098–7099 (1976).
Sternson, L. A., Stobaugh, J. F. & Repta, A. J. Rational design and evaluation of improved o-phthalaldehyde-like fluorogenic reagents. Anal. Biochem. 144, 233–246 (1985).
Jacobson, S. C., Koutny, L. B., Hergenroeder, R., Moore, A. W. & Ramsey, J. M. Microchip capillary electrophoresis with an integrated postcolumn reactor. Anal. Chem. 66, 3472–3476 (1994).
Otzen, D. Protein–surfactant interactions: a tale of many states. Biochim. Biophys. Acta 1814, 562–591 (2011).
Stobaugh, J., Repta, A., Sternson, L. & Garren, K. Factors affecting the stability of fluorescent isoindoles derived from reaction of o-phthalaldehyde and hydroxyalkylthiols with primary amines. Anal. Biochem. 135, 495–504 (1983).
Lindroth, P. & Mopper, K. High performance liquid chromatographic determination of subpicomole amounts of amino acids by precolumn fluorescence derivatization with o-phthaldialdehyde. Anal. Chem. 51, 1667–1674 (1979).
Svedas, V. J., Galaev, I. J., Borisov, I. L. & Berezin, I. V. The interaction of amino acids with o-phthalaldehyde: a kinetic study and spectrophometric assay of the reaction product. Anal. Biochem. 101, 188–195 (1980).
Horrocks, M. H. et al. Single-molecule measurements of transient biomolecular complexes through microfluidic dilution. Anal. Chem. 85, 6855–6859 (2013).
Waters, J. C. Accuracy and precision in quantitative fluorescence microscopy. J. Cell Biol. 185, 1135–1148 (2009).
De Genst, E. J. et al. Structure and properties of a complex of alpha-synuclein and a single-domain camelid antibody. J. Mol. Biol. 402, 326–343 (2010).
Jha, A. K., Colubri, A., Freed, K. F. & Sosnick, T. R. Statistical coil model of the unfolded state: resolving the reconciliation problem. Proc. Natl Acad. Sci. USA, 102, 13099–13104 (2005).
Ulmer, T. S., Bax, A., Cole, N. B. & Nussbaum, R. L. Structure and dynamics of micelle-bound human α-synuclein. J. Biol. Chem. 280, 9595–9603 (2005).
Dedmon, M. M., Lindorff-Larsen, K., Christodoulou, J., Vendruscolo, M. & Dobson, C. M. Mapping long-range interactions in α-Synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 127, 476–477 (2005).
Morar, A. S., Olteanu, A., Young, G. B. & Pielak, G. J. Solvent-induced collapse of α-synuclein and acid-denatured cytochrome c. Prot. Sci. 10, 2195–2199 (2001).
Weinreb, P. H., Zhen, W., Poon, A. W., Conway, K. A. & Lansbury, P. T. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry 35, 13709–13715 (1996).
Lord, R. S., Gubensek, F. & Rupley, J. A. Insulin self-association. Spectrum changes and thermodynamics. Biochemistry 12, 4385–4392 (1973).
Bocian, W. et al. Structure of human insulin monomer in water/acetonitrile solution. J. Biomol. NMR 40, 55–64 (2008).
Lin, M. & Larive, C. Detection of insulin aggregates with pulsed-field gradient nuclear magnetic resonance spectroscopy. Anal. Biochem. 229, 214–220 (1995).
Muyldermans, S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797 (2013).
De Genst, E., Messer, A. & Dobson, C. M. Antibodies and protein misfolding: from structural research tools to therapeutic strategies. Biochim. Biophys. Acta 1844, 1907–1919 (2014).
De Genst, E. & Dobson, C. M. in Methods in Molecular Biology Vol. 911 (eds Saerens, D. & Mulydermans, S.), 533–558 (2012).
Guilliams, T. et al. Nanobodies raised against monomeric α-synuclein distinguish between fibrils at different maturation stages. J. Mol. Biol. 425, 2397–2411 (2013).
Acknowledgements
The authors acknowledge the European Research Council, Biotechnology and Biological Sciences Research Council, Wellcome Trust, Newman Foundation, Winston Churchill Foundation and Elan Pharmaceuticals for financial support. E.D.G. was supported by the Medical Research Council (G1002272). The authors thank J. Steyaert at the Free University of Brussels for sharing the NbSyn1a clone.
Author information
Authors and Affiliations
Contributions
T.P.J.K. and C.M.D. supervised the research. E.V.Y., L.R., M.V., C.M.D. and T.P.J.K. conceived and designed the experiments. E.V.Y. performed the experiments. E.V.Y. and T.M. analysed the data. E.J.D.G., P.A. and S.L. contributed materials and/or analysis tools. E.V.Y., C.M.D. and T.P.J.K. wrote the paper, and all authors commented on the paper.
Corresponding authors
Ethics declarations
Competing interests
Part of the work described here has been the subject of a patent application filed by Cambridge Enterprise Ltd, a fully owned subsidiary of the University of Cambridge (now licensed to Fluidic Analytics, of which C.M.D. is a scientific advisor and T.P.J.K. is a board member).
Supplementary information
Supplementary information
Supplementary information (PDF 821 kb)
Rights and permissions
About this article
Cite this article
Yates, E., Müller, T., Rajah, L. et al. Latent analysis of unmodified biomolecules and their complexes in solution with attomole detection sensitivity. Nature Chem 7, 802–809 (2015). https://doi.org/10.1038/nchem.2344
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2344
This article is cited by
-
Widespread amyloidogenicity potential of multiple myeloma patient-derived immunoglobulin light chains
BMC Biology (2023)
-
On the utility of microfluidic systems to study protein interactions: advantages, challenges, and applications
European Biophysics Journal (2023)
-
Quantifying misfolded protein oligomers as drug targets and biomarkers in Alzheimer and Parkinson diseases
Nature Reviews Chemistry (2021)
-
Kinetic fingerprints differentiate the mechanisms of action of anti-Aβ antibodies
Nature Structural & Molecular Biology (2020)
-
Microfluidic approaches for the analysis of protein–protein interactions in solution
Biophysical Reviews (2020)