Abstract
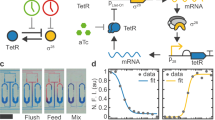
In vitro compartmentalization of biochemical reaction networks is a crucial step towards engineering artificial cell-scale devices and systems. At this scale the dynamics of molecular systems becomes stochastic, which introduces several engineering challenges and opportunities. Here we study a programmable transcriptional oscillator system that is compartmentalized into microemulsion droplets with volumes between 33 fl and 16 pl. Simultaneous measurement of large populations of droplets reveals major variations in the amplitude, frequency and damping of the oscillations. Variability increases for smaller droplets and depends on the operating point of the oscillator. Rather than reflecting the stochastic kinetics of the chemical reaction network itself, the variability can be attributed to the statistical variation of reactant concentrations created during their partitioning into droplets. We anticipate that robustness to partitioning variability will be a critical challenge for engineering cell-scale systems, and that highly parallel time-series acquisition from microemulsion droplets will become a key tool for characterization of stochastic circuit function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
22 April 2014
The authors wish to add the following to the Acknowledgements section of this Article ‘E.F. and F.C.S. gratefully acknowledge funding from the Bavaria California Technology Center (BaCaTeC).’ The online versions of the Article have been amended accordingly.
References
Kim, J., White, K. S. & Winfree, E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Sys. Biol. 2, 68 (2006).
Forster, A. C. & Church, G. M. Synthetic biology projects in vitro. Genome Res. 17, 1–6 (2007).
Montagne, K., Plasson, R., Sakai, Y., Fujii, T. & Rondelez, Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466 (2011).
Zhou, H-X., Rivas, G. & Minton, A. P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Ann. Rev. Biophys. 37, 375–397 (2008).
De, T. & Maitra, A. Solution behaviour of aerosol OT in non-polar solvents. Adv. Colloid Interface Sci. 59, 95–193 (1995).
Gillespie, D. T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977).
Noireaux, V. & Libchaber, A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl Acad. Sci. USA 101, 17669–17674 (2004).
Shin, J. & Noireaux, V. An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synthetic Biol. 1, 29–41 (2012).
Matsuura, T. et al. Effects of compartment size on the kinetics of intracompartmental multimeric protein synthesis. ACS Synthetic Biol. 1, 431–437 (2012).
Theberge, A. B. et al. Microdroplets in microfluidics: an evolving platform for discoveries in chemistry and biology. Angew. Chem. Int. Ed. 49, 5846–5868 (2010).
Rotman, B. Measurement of activity of single molecules of beta-D-galactosidase. Proc. Natl Acad. Sci. USA 47, 1981–1991 (1961).
Nakano, M. et al. Single-molecule PCR using water-in-oil emulsion. J. Biotechnol. 102, 117–124 (2003).
Miller, O. J. et al. Directed evolution by in vitro compartmentalization. Nature Methods 3, 561–570 (2006).
Lu, W-C. & Ellington, A. D. In vitro selection of proteins via emulsion compartments. Methods 60, 75–80 (2013).
Epstein, I. et al. Chemical oscillators in structured media. Acc. Chem. Res. 45, 2160–2168 (2011).
Nagypal, I. & Epstein, I. R. Fluctuations and stirring rate effects in the chlorite thiosulfate reaction. J. Phys. Chem. 90, 6285–6292 (1986).
Vanag, V. & Epstein, I. Pattern formation in a tunable medium: the Belousov–Zhabotinsky reaction in an aerosol OT microemulsion. Phys. Rev. Lett. 87, 228301 (2001).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Fung, E. et al. A synthetic gene-metabolic oscillator. Nature 435, 118–122 (2005).
Stricker, J. et al. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–539 (2008).
Tigges, M., Marquez-Lago, T. T., Stelling, J. & Fussenegger, M. A tunable synthetic mammalian oscillator. Nature 457, 309–312 (2009).
Danino, T., Mondragón-Palomino, O., Tsimring, L. & Hasty, J. A synchronized quorum of genetic clocks. Nature 463, 326–330 (2010).
Ackermann, J., Wlotzka, B. & McCaskill, J. In vitro DNA-based predator–prey system with oscillatory kinetics. Bull. Math. Biol. 60, 329–354 (1998).
Kim, J. & Winfree, E. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 7, 465 (2011).
Franco, E. et al. Timing molecular motion and production with a synthetic transcriptional clock. Proc. Natl Acad. Sci. USA 108, E784–E793 (2011).
Winfree, A. T. The Geometry of Biological Time (Springer, 1980).
Gonze, D., Halloy, J. & Goldbeter, A. Robustness of circadian rhythms with respect to molecular noise. Proc. Natl Acad. Sci. USA 99, 673–678 (2002).
Vilar, J. M. G., Kueh, H. Y., Barkai, N. & Leibler, S. Mechanisms of noise-resistance in genetic oscillators. Proc. Natl Acad. Sci. USA 99, 5988–5992 (2002).
Kar, S., Baumann, W. T., Paul, M. R. & Tyson, J. J. Exploring the roles of noise in the eukaryotic cell cycle. Proc. Natl Acad. Sci. USA 106, 6471–6476 (2009).
Hong, C. I., Conrad, E. D. & Tyson, J. J. A proposal for robust temperature compensation of circadian rhythms. Proc. Natl Acad. Sci. USA. 104, 1195–1200 (2007).
Fujii, T. & Rondelez, Y. Predator–prey molecular ecosystems. ACS Nano 7, 27–34 (2013).
Hasatani, K. et al. High-throughput and long-term observation of compartmentalized biochemical oscillators. Chem. Comm. 49, 8090–8092 (2013).
Kaern, M., Elston, T. C., Blake, W. J. & Collins, J. J. Stochasticity in gene expression: from theories to phenotypes. Nature Rev. Genet. 6, 451–464 (2005).
Raj, A. & van Oudenaarden, A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008).
Huh, D. & Paulsson, J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nature Genet. 43, 95–100 (2011).
Huh, D. & Paulsson, J. Random partitioning of molecules at cell division. Proc. Natl Acad. Sci. USA 108, 15004–15009 (2011).
Rizzo, J., Gifford, L. K., Zhang, X., Gewirtz, A. M. & Lu, P. Chimeric RNA–DNA molecular beacon assay for ribonuclease H activity. 16, 277–283 (2002).
Holtze, C. et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab on a Chip 8, 1632–1639 (2008).
Liu, Y., Jung, S-Y. & Collier, C. P. Shear-driven redistribution of surfactant affects enzyme activity in well-mixed femtoliter droplets. Anal. Chem. 81, 4922–4928 (2009).
Maslak, M. & Martin, C. T. Effects of solution conditions on the steady-state kinetics of initiation of transcription by T7 RNA polymerase. Biochemistry 33, 6918–6924 (1994).
Friedman, N., Cai, L. & Xie, X. S. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys. Rev. Lett. 97 (2006).
Modesti, M. in Single Molecule Analysis (eds Peterman, E. J. G. & Wuite, G. J. L.) 101–120 (Methods in Molecular Biology 783, Humana Press, 2011).
Gutenkunst, R. N. et al. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 3, e189 (2007).
Subsoontorn, P., Kim, J. & Winfree, E. Ensemble Bayesian analysis of bistability in a synthetic transcriptional switch. ACS Synthetic Biol. 1, 299–316 (2012).
Schütze, T. et al. A streamlined protocol for emulsion polymerase chain reaction and subsequent purification. Anal. Biochem. 410, 155–157 (2011).
Acknowledgements
The authors acknowledge financial support by the National Science Foundation grants CCF-0832824 (The Molecular Programming Project) and CMMI-1266402, by the Bourns College of Engineering at the University of California at Riverside (UC), the UC Regents Faculty Development Fellowship, the European Commission FP7 grant no. 248919 (Bacterial Computing with Engineered Populations), the German Research Foundation Cluster of Excellence Nanosystems Initiative Munich and the Elite Network of Bayern. Surfactant E2K0660 was supplied by RainDance Technologies. We acknowledge E. Friedrichs and R. Jungmann for initial experiments; U. Gerland, R. Murray and N. Karlsson for useful discussions, advice and support; and C. Martin, L. Ramìrez-Tapia, E. Stahl and H. Dietz for providing fluorescently labelled T7 RNAP.
Author information
Authors and Affiliations
Contributions
E.W., E.F. and F.C.S. designed the research; M.W., J.K. and E.F. performed the research; M.W., J.K., K.K. and E.F. analysed the data; E.W., E.F. and F.C.S wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 23789 kb)
Supplementary Movie 1
Supplementary Movie 1 (MOV 2854 kb)
Supplementary Movie 2
Supplementary Movie 2 (MOV 2223 kb)
Supplementary Movie 3
Supplementary Movie 3 (MOV 4665 kb)
Rights and permissions
About this article
Cite this article
Weitz, M., Kim, J., Kapsner, K. et al. Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nature Chem 6, 295–302 (2014). https://doi.org/10.1038/nchem.1869
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1869
This article is cited by
-
A genetic circuit on a single DNA molecule as an autonomous dissipative nanodevice
Nature Communications (2024)
-
Superstructural ordering in self-sorting coacervate-based protocell networks
Nature Chemistry (2024)
-
An oscillating reaction network with an exact closed form solution in the time domain
BMC Bioinformatics (2023)
-
Stochastic approximations of higher-molecular by bi-molecular reactions
Journal of Mathematical Biology (2023)
-
Programmable synthetic cell networks regulated by tuneable reaction rates
Nature Communications (2022)