Abstract
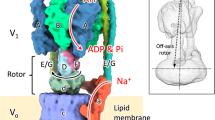
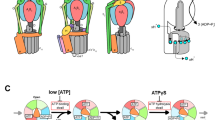
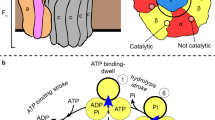
Rotary ATPases play fundamental roles in energy conversion as their catalytic rotation is associated with interdomain fluctuations and heterogeneity of conformational states. Using ion mobility mass spectrometry we compared the conformational dynamics of the intact ATPase from Thermus thermophilus with those of its membrane and soluble subcomplexes. Our results define regions with enhanced flexibility assigned to distinct subunits within the overall assembly. To provide a structural context for our experimental data we performed molecular dynamics simulations and observed conformational changes of the peripheral stalks that reflect their intrinsic flexibility. By isolating complexes at different phases of cell growth and manipulating nucleotides, metal ions and pH during isolation, we reveal differences that can be related to conformational changes in the Vo complex triggered by ATP binding. Together these results implicate nucleotides in modulating flexibility of the stator components and uncover mechanistic detail that underlies operation and regulation in the context of the holoenzyme.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
von Ballmoos, C., Cook, G. M. & Dimroth, P. Unique rotary ATP synthase and its biological diversity. Annu. Rev. Biophys. 37, 43–64 (2008).
Stewart, A. G., Sobti, M., Harvey, R. P. & Stock, D. Rotary ATPases: models, machine elements and technical specifications. Bioarchitecture 3, 2–12 (2013).
Nakano, M. et al. ATP hydrolysis and synthesis of a rotary motor V-ATPase from Thermus thermophilus. J. Biol. Chem. 283, 20789–20796 (2008).
Feniouk, B. A., Suzuki, T & Yoshida, M. Regulatory interplay between proton motive force, ADP, phosphate, and subunit epsilon in bacterial ATP synthase. J. Biol. Chem. 282, 764–772 (2007).
Muench, S. P., Trinick, J. & Harrison, M. A. Structural divergence of the rotary ATPases. Q. Rev. Biophys. 44, 311–356 (2011).
Oot, R. A., Huang, L. S., Berry, E. A. & Wilkens, S. Crystal structure of the yeast vacuolar ATPase heterotrimeric EGC(head) peripheral stalk complex. Structure 20, 1881–1892 (2012).
Yokoyama, K. et al. Subunit arrangement in V-ATPase from Thermus thermophilus. J. Biol. Chem. 278, 42686–42691 (2003).
Barrera, N. P. et al. Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nature Methods 6, 585–587 (2009).
Barrera, N. P., Di Bartolo, N., Booth, P. J. & Robinson, C. V. Micelles protect membrane complexes from solution to vacuum. Science 321, 243–246 (2008).
Wang, S. C. et al. Ion mobility mass spectrometry of two tetrameric membrane protein complexes reveals compact structures and differences in stability and packing. J. Am. Chem. Soc. 132, 15468–15470 (2010).
Borysik, A. J., Hewitt, D. J. & Robinson, C. V. Detergent release prolongs the lifetime of native-like membrane protein conformations in the gas-phase. J. Am. Chem. Soc. 135, 6078–6083 (2013).
Bohrer, B. C., Merenbloom, S. I., Koeniger, S. L., Hilderbrand, A. E. & Clemmer, D. E. Biomolecule analysis by ion mobility spectrometry. Annu. Rev. Anal. Chem. 1, 293–327 (2008).
Bleiholder, C., Dupuis, N. F., Wyttenbach, T. & Bowers, M. T. Ion mobility–mass spectrometry reveals a conformational conversion from random assembly to beta-sheet in amyloid fibril formation. Nature Chem. 3, 172–177 (2011).
Grabenauer, M. et al. Conformational stability of Syrian hamster prion protein PrP(90–231). J. Am. Chem. Soc. 132 (26), 8816–8818 (2010).
Dupuis, N. F., Wu, C., Shea, J. E. & Bowers, M. T. Human islet amyloid polypeptide monomers form ordered beta-hairpins: a possible direct amyloidogenic precursor. J. Am. Chem. Soc. 131, 18283–18292 (2009).
Michaelevski, I., Kirshenbaum, N. & Sharon, M. T-wave ion mobility–mass spectrometry: basic experimental procedures for protein complex analysis. J. Vis. Exp. 41, e1985, doi:10.3791/1985 (2010).
Ruotolo, B. T., Benesch, J. L., Sandercock, A. M., Hyung, S. J. & Robinson, C. V. Ion mobility–mass spectrometry analysis of large protein complexes. Nature Protocols 3, 1139–1152 (2008).
Hall, Z., Politis, A. & Robinson, C. V. Structural modeling of heteromeric protein complexes from disassembly pathways and ion mobility–mass spectrometry. Structure 20, 1596–1609 (2012).
Alber, F. et al. Determining the architectures of macromolecular assemblies. Nature 450, 683–694 (2007).
Hall, Z., Politis, A., Bush, M. F., Smith, L. J. & Robinson, C. V. Charge-state dependent compaction and dissociation of protein complexes: insights from ion mobility and molecular dynamics. J. Am. Chem. Soc. 134, 3429–3438 (2012).
Zhou, M. et al. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science 334, 380–385 (2011).
Barrera, N. P., Zhou, M. & Robinson, C. V. The role of lipids in defining membrane protein interactions: insights from mass spectrometry. Trends Cell. Biol. 23, 1–8 (2013).
Morgner, N., Montenegro, F., Barrera, N. P. & Robinson, C. V. Mass spectrometry – from peripheral proteins to membrane motors. J. Mol. Biol. 423, 1–13 (2012).
Bush, M. F. et al. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal. Chem. 82, 9557–9565 (2010).
Zhong, Y., Hyung, S. J. & Ruotolo, B. T. Characterizing the resolution and accuracy of a second-generation traveling-wave ion mobility separator for biomolecular ions. Analyst 136, 3534–3541 (2011).
Shvartsburg, A. A. & Smith, R. D. Fundamentals of traveling wave ion mobility spectrometry. Anal. Chem. 80, 9689–9699 (2008).
Giles, K. et al. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid. Commun. Mass Spectrom. 18, 2401–2414 (2004).
Nagamatsu, Y., Takeda, K., Kuranaga, T., Numoto, N. & Miki, K. Origin of asymmetry at the intersubunit interfaces of V1-ATPase from Thermus thermophilus. J. Mol. Biol. 425, 2699–2708 (2013).
Suhre, K. & Sanejouand, Y. H. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 32, W610–W614 (2004).
Drory, O., Frolow, F. & Nelson, N. Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO Rep. 5, 1148–1152 (2004).
Lee, L. K., Stewart, A. G., Donohoe, M., Bernal, R. A. & Stock, D. The structure of the peripheral stalk of Thermus thermophilus H+-ATPase/synthase. Nature Struct. Mol. Biol. 17, 373–378 (2010).
Bernal, R. A. & Stock, D. Three-dimensional structure of the intact Thermus thermophilus H+-ATPase/synthase by electron microscopy. Structure 12, 1789–1798 (2004).
Stewart, A. G., Lee, L. K., Donohoe, M., Chaston, J. J. & Stock, D. The dynamic stator stalk of rotary ATPases. Nature Commun. 3, 687 (2012).
Kinosita, K. Jr, Yasuda, R., Noji, H. & Adachi, K. A rotary molecular motor that can work at near 100% efficiency. Philos. Trans. R. Soc. Lond. B 355, 473–489 (2000).
Esteban, O., Christ, D. & Stock, D. Purification of molecular machines and nanomotors using phage-derived monoclonal antibody fragments. Methods Mol. Biol. 996, 203–217 (2013).
Arai, S. et al. Rotation mechanism of Enterococcus hirae V1-ATPase based on asymmetric crystal structures. Nature 493, 703–707 (2013).
Giles, K., Williams, J. P. & Campuzano, I. Enhancements in travelling wave ion mobility resolution. Rapid. Commun. Mass Spectrom. 25, 1559–1566 (2011).
Pagel, K., Natan, E., Hall, Z., Fersht, A. R. & Robinson, C. V. Intrinsically disordered p53 and its complexes populate compact conformations in the gas phase. Angew. Chem. Int. Ed. 52, 361–365 (2013).
Armbruster, A. et al. Evidence for major structural changes in subunit C of the vacuolar ATPase due to nucleotide binding. FEBS Lett. 579, 1961–1967 (2005).
Lau, W. C. & Rubinstein, J. L. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature 481, 214–218 (2012).
Beis, I. & Newsholme, E. A. The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates. Biochem. J. 152, 23–32 (1975).
Schneider, D. A. & Gourse, R. L. Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J. Biol. Chem. 279, 8262–8268 (2004).
Tani, K. et al. Visualization of two distinct states of disassembly in the bacterial V-ATPase from Thermus thermophilus. Microscopy 62 (4), 467–474 (2013).
Armbruster, A. et al. Structural analysis of the stalk subunit Vma5p of the yeast V-ATPase in solution. FEBS Lett. 570, 119–125 (2004).
Hernandez, H. & Robinson, C. V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature Protocols 2, 715–726 (2007).
van der Spoel, D., et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Reimann, C. T., Velazquez, I. & Tapia, O. Proteins in vacuo. Denaturation of highly-charged lysozyme studied by molecular dynamics simulations. J. Phys. Chem. B 102, 9344–9352 (1998).
Acknowledgements
This work was funded by the Wellcome Trust (M.Z. and C.V.R.) and by a PROSPECTS (HEALTHF4-2008-201648) grant within the Research Framework of the European Union (A.P. and C.V.R.), together with Engineering and Physical Sciences Research Council funding for research scientists with caring responsibilities (M.Z.), funding from the Royal Society and an European Research Council investigator award Integral Membrane Proteins Resolution of Stoichiometry and Structure (IMPRESS) (C.V.R.). D.S., R.B.D. and A.G.S. are funded by Australian National Health and Medical Research Council grants 1004620, 1022143 and 1047004, respectively.
Author information
Authors and Affiliations
Contributions
C.V.R. and D.S designed the research, M.Z. conducted all IM–MS experiments and analysed the data, R.B.D. purified TtATPases, A.P. and A.G.S. performed the modelling, A.P. conducted MD simulations and all CCS calculations, I.L. carried out the MS analysis of dAb:TtATPase binding, K.J.W. conducted the Western blot experiments, and M.Z. and C.V.R. and A.P. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 2158 kb)
Supplementary Movie 1
Supplementary Movie 1 (AVI 12413 kb)
Supplementary Movie 2
Supplementary Movie 2 (AVI 9252 kb)
Rights and permissions
About this article
Cite this article
Zhou, M., Politis, A., Davies, R. et al. Ion mobility–mass spectrometry of a rotary ATPase reveals ATP-induced reduction in conformational flexibility. Nature Chem 6, 208–215 (2014). https://doi.org/10.1038/nchem.1868
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1868
This article is cited by
-
Cross-linking mass spectrometry uncovers protein interactions and functional assemblies in synaptic vesicle membranes
Nature Communications (2021)
-
Reconstitution of FoF1-ATPase-based biomimetic systems
Nature Reviews Chemistry (2019)
-
Integrating hydrogen–deuterium exchange mass spectrometry with molecular dynamics simulations to probe lipid-modulated conformational changes in membrane proteins
Nature Protocols (2019)
-
Characterization of Conformational Ensembles of Protonated N-glycans in the Gas-Phase
Scientific Reports (2018)
-
Native Top-Down Mass Spectrometry and Ion Mobility MS for Characterizing the Cobalt and Manganese Metal Binding of α-Synuclein Protein
Journal of the American Society for Mass Spectrometry (2018)