Abstract
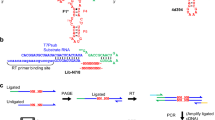
Vitamins are hypothesized to be relics of an RNA world, and were probably participants in RNA-mediated primordial metabolism. If catalytic RNAs, or ribozymes, could harness vitamin cofactors to aid their function in a manner similar to protein enzymes, it would enable them to catalyse a much larger set of chemical reactions. The cofactor thiamin diphosphate, a derivative of vitamin B1 (thiamin), is used by enzymes to catalyse difficult metabolic reactions, including decarboxylation of stable α-keto acids such as pyruvate. Here, we report a ribozyme that uses free thiamin to decarboxylate a pyruvate-based suicide substrate (LnkPB). Thiamin conjugated to biotin was used to isolate catalytic individuals from a pool of random-sequence RNAs attached to LnkPB. Analysis of a stable guanosine adduct obtained via digestion of an RNA sequence (clone dc4) showed the expected decarboxylation product. The discovery of a prototypic thiamin-utilizing ribozyme has implications for the role of RNA in orchestrating early metabolic cycles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kruger, K. et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157 (1982).
Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857 (1983).
White, H. B. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 7, 101–104 (1975).
Gilbert, W. Origin of life: the RNA world. Nature 319, 618 (1986).
Benner, S. A., Ellington, A. D. & Tauer, A. Modern metabolism as a palimpsest of the RNA world. Proc. Natl Acad. Sci. USA 86, 7654–7656 (1989).
Tucker, B. J. & Breaker, R. R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 15, 342–348 (2005).
Marchfelder, A. et al. Small RNAs for defence and regulation in Archaea. Extremophiles 16, 685–696 (2012).
Irnov, I., Sharma, C. M., Vogel, J. & Winkler, W. C. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 38, 6637–6651 (2010).
Durand, S. & Storz, G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 75, 1215–1231 (2010).
Liu, J. M. & Camilli, A. A broadening world of bacterial small RNAs. Curr. Opin. Microbiol. 13, 18–23 (2010).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Robertson, D. L. & Joyce, G. F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344, 467–468 (1990).
Winkler, W. C., Nahvi, A., Roth, A., Collins, J. A. & Breaker, R. R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004).
Jadhav, V. R. & Yarus, M. Coenzymes as coribozymes. Biochimie 84, 877–888 (2002).
Tsukiji, S., Pattnaik, S. B. & Suga, H. An alcohol dehydrogenase ribozyme. Nature Struct. Mol. Biol. 10, 713–717 (2004).
Tsukiji, S., Pattnaik, S. B. & Suga, H. Reduction of an aldehyde by a NADH/Zn2+-dependent redox active ribozyme dehydrogenase ribozyme. J. Am. Chem. Soc. 126, 5044–5045 (2004).
Jadhav, V. R. & Yarus, M. Acyl-CoAs from coenzyme ribozymes. Biochemistry 41, 723–729 (2002).
Li, N. & Huang, F. Ribozyme-catalyzed aminoacylation from CoA thioesters. Biochemistry 44, 4582–4590 (2002).
Coleman, T. M. & Huang, F. RNA-catalyzed thioester synthesis. Chem. Biol. 9, 1227–1236 (2002).
Kluger, R. & Tittmann, K. Thiamin diphosphate catalysis: enzymic & nonenzymic covalent intermediates. Chem. Rev. 108, 1797–1833 (2008).
Walsh, C. T. Suicide substrates, mechanism-based enzyme inactivators: recent developments. Ann. Rev. Biochem. 53, 493–535 (1984).
Jordan, F. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat. Prod. Rep. 20, 184–201 (2003).
Schellenberger, A. Sixty years of thiamin diphosphate biochemistry. Biochim. Biophys. Acta 1385, 177–186 (1998).
Breslow, R. On the mechanism of thiamin action. IV. Evidence from studies on model systems. J. Am. Chem. Soc. 80, 3719–3726 (1958).
Kuo, D. J. & Jordan, F. Active site directed irreversible inactivation of brewers’ yeast pyruvate decarboxylase by the conjugated substrate analogue (E)-4-(4-chlorophenyl)-2-oxo-3-butenoic acid: development of a suicide substrate. Biochemistry 22, 3735–3740 (1983).
Kuo, D. J. & Jordan, F. Direct spectroscopic observation of a brewer's yeast pyruvate decarboxylase-bound enamine intermediate produced from a suicide substrate. J. Biol. Chem. 258, 13415–13417 (1983).
Kluger, R., Chin, J. & Smyth, T. Thiamin-catalyzed decarboxylation of pyruvate. Synthesis and reactivity analysis of the central, elusive intermediate, α-lactylthiamin J. Am. Chem. Soc. 103, 884–888 (1981).
Washabaugh, M. W. & Jencks, W. P. Thiazolium C(2)-proton exchange: general-base catalysis, direct proton transfer, and acid inhibition. Biochemistry 27, 5044–5053 (1988).
Cheong, C., Varani, G. & Tinoco, I. Jr. Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature 346, 680–682 (1990).
Doudna, J. A. & Cech, T. R. The chemical repertoire of natural ribozymes. Nature 418, 222–228 (2002).
Chandrasekar, J. & Silverman, S. K. Catalytic DNA with phosphatase activity. Proc. Natl Acad. Sci. USA 110, 5315–5320 (2013).
Kemp, D. S. & O'Brien, J. T. Base catalysis of thiazolium salt hydrogen exchange and its implications for enzymic thiamine cofactor catalysis. J. Am. Chem. Soc. 92, 2554–2555 (1970).
Zhang, S. et al. Evidence for dramatic acceleration of a C–H bond ionization rate in thiamin diphosphate enzymes by the protein environment. Biochemistry 44, 2237–2243 (2005).
Frank, R. A., Leeper, F. J. & Luisi, B. F. Structure, mechanism and catalytic duality of thiamin-dependent enzymes. Cell. Mol. Life Sci. 64, 892–905 (2007).
Kraut, J. & Reed, H. J. The crystal structure of thiamin hydrochloride (vitamin B1). Acta Crystallogr. 15, 747–749 (1962).
Jordan, F., Li, H. & Brown, A. Remarkable stabilization of zwitterionic intermediates may account for a billion-fold rate acceleration by thiamin diphosphate-dependent decarboxylases. Biochemistry 38, 6369–6373 (1999).
Jordan, F. et al. Dual catalytic apparatus of the thiamin diphosphate coenzyme: acid–base via the 1′,4′-iminopyrimidine tautomer along with its electrophilic role. J. Am. Chem. Soc. 125, 12732–12738 (2003).
Kern, D. et al. How thiamin diphosphate is activated in enzymes. Science 275, 67–70 (1997).
Winkler, W. C., Nahvi, A. & Breaker, R. R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002).
Cochrane, J. C. & Strobel, S. A. Riboswitch effectors as protein enzyme cofactors. RNA 14, 993–1002 (2008).
Thore, S., Leibundgut, M. & Ban, N. Structure of the eukaryotic thiamin pyrophosphate riboswitch with its regulatory ligand. Science 312, 1208–1211 (2006).
Serganov, A., Polonskaia, A., Phan, A. T., Breaker, R. & Patel, D. J. Structural basis for gene regulation by a thiamin pyrophosphate-sensing riboswitch. Nature 441, 1167–1171 (2006).
Edwards, T. & Ferré-D'Amaré, A. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA–small molecule recognition. Structure 14, 1459–1468 (2006).
Acknowledgements
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC). The authors thank R. N. Young and the members of his group for advice and assistance with organic synthesis, and N. S. Kumar, A. J. Bennet, P. J. Unrau, E. C. Young, A. R. Lewis and H. Chen for their expert and practical advice.
Author information
Authors and Affiliations
Contributions
P.C. and D.S. conceived and designed the experiments. P.C. performed the experiments. P.C. and D.S. analysed the data. P.C. and D.S. co-wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 677 kb)
Rights and permissions
About this article
Cite this article
Cernak, P., Sen, D. A thiamin-utilizing ribozyme decarboxylates a pyruvate-like substrate. Nature Chem 5, 971–977 (2013). https://doi.org/10.1038/nchem.1777
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1777
This article is cited by
-
Structure and mechanism of the methyltransferase ribozyme MTR1
Nature Chemical Biology (2022)
-
The GlcN6P cofactor plays multiple catalytic roles in the glmS ribozyme
Nature Chemical Biology (2017)
-
Ribozyme takes its vitamins
Nature Chemistry (2013)