Abstract
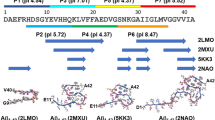
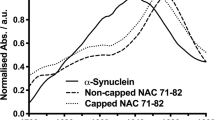
The amyloid protein aggregation associated with diseases such as Alzheimer's, Parkinson's and type II diabetes (among many others) features a bewildering variety of β-sheet-rich structures in transition from native proteins to ordered oligomers and fibres. The variation in the amino-acid sequences of the β-structures presents a challenge to developing a model system of β-sheets for the study of various amyloid aggregates. Here, we introduce a family of robust β-sheet macrocycles that can serve as a platform to display a variety of heptapeptide sequences from different amyloid proteins. We have tailored these amyloid β-sheet mimics (ABSMs) to antagonize the aggregation of various amyloid proteins, thereby reducing the toxicity of amyloid aggregates. We describe the structures and inhibitory properties of ABSMs containing amyloidogenic peptides from the amyloid-β peptide associated with Alzheimer's disease, β2-microglobulin associated with dialysis-related amyloidosis, α-synuclein associated with Parkinson's disease, islet amyloid polypeptide associated with type II diabetes, human and yeast prion proteins, and Tau, which forms neurofibrillary tangles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Aguzzi, A. & O'Connor, T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nature Rev. Drug. Discov. 9, 237–248 (2010).
Bartolini, M. & Andrisano, V. Strategies for the inhibition of protein aggregation in human diseases. ChemBioChem 11, 1018–1035 (2010).
Greenwald, J. & Riek, R. Biology of amyloid: structure, function, and regulation. Structure 18, 1244–1260 (2010).
Tycko, R. Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem. 62, 279–299 (2011).
Eichner, T. & Radford, S. E. A diversity of assembly mechanisms of a generic amyloid fold. Mol. Cell 43, 8–18 (2011).
Lopez de la Paz, M. & Serrano, L. Sequence determinants of amyloid fibril formation. Proc. Natl Acad. Sci. USA 101, 87–92 (2004).
Goldschmidt, L., Teng, P. K., Riek, R. & Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl Acad. Sci. USA 107, 3487–3492 (2010).
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005).
Sawaya, M. R. et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447, 453–457 (2007).
Conway, K. A. et al. Acceleration of oligomerization, not fibrillization, is a shared property of both α-synuclein mutations linked to early-onset Parkinson's disease: implications for pathogenesis and therapy. Proc. Natl Acad. Sci. USA 97, 571–576 (2000).
Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T. & Lansbury, P. T. Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature 418, 291–291 (2002).
Chimon, S. et al. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer's β-amyloid. Nature Struct. Mol. Biol. 14, 1157–1164 (2007).
Bernstein, S. L. et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nature Chem. 1, 326–331 (2009).
Ono, K., Condron, M. M. & Teplow D. B. Structure–neurotoxicity relationships of amyloid β-protein oligomers. Proc. Natl Acad. Sci. USA 106, 14745–14750 (2009).
Woods, R. J. et al. Cyclic modular β-sheets. J. Am. Chem. Soc. 129, 2548–2558 (2007).
Liu, C. et al. Characteristics of amyloid-related oligomers revealed by crystal structures of macrocyclic β-sheet mimics. J. Am. Chem. Soc. 133, 6736–6744 (2011).
Zheng, J. et al. Macrocyclic β-sheet peptides that inhibit the aggregation of a tau-protein-derived hexapeptide. J. Am. Chem. Soc. 133, 3144–3157 (2011).
Gellman, S. H. Minimal model systems for β-sheet secondary structure in proteins. Curr. Opin. Chem. Biol. 2, 717–725 (1998).
Nowick, J. S. et al. An unnatural amino acid that mimics a tripeptide β-strand and forms β-sheetlike hydrogen-bonded dimers. J. Am. Chem. Soc. 122, 7654–7661 (2000).
Nowick, J. S. & Brower, J. O. A new turn structure for the formation of β-hairpins in peptides. J. Am. Chem. Soc. 125, 876–877 (2003).
Finder, V. H. & Glockshuber, R. Amyloid-β aggregation. Neurodegen. Dis. 4, 13–27 (2007).
Walsh, P., Simonetti, K. & Sharpe, S. Core structure of amyloid fibrils formed by residues 106–126 of the human prion protein. Structure 17, 417–426 (2009).
Friedhoff, P., von Bergen, M., Mandelkow, E-M., Davies, P. & Mandelkow, E. A nucleated assembly mechanism of Alzheimer paired helical filaments. Proc. Natl Acad. Sci. USA 95, 15712–15717 (1998).
Platt, G. W., Routledge, K. E., Homans, S. W. & Radford, S. E. Fibril growth kinetics reveal a region of β2-microglobulin important for nucleation and elongation of aggregation. J. Mol. Biol. 378, 251–263 (2008).
Vilar, M. et al. The fold of α-synuclein fibrils. Proc. Natl Acad. Sci. USA 105, 8637–8642 (2008).
Luca, S., Yau, W-M., Leapman, R. & Tycko, R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry 46, 13505–13522 (2007).
Cheng, P-N. & Nowick, J. S. Giant macrolactams based on β-sheet peptides. J. Org. Chem. 76, 3166–3173 (2011).
Yan, L-M., Velkova, A., Tatarek-Nossol, M., Andreetto, E. & Kapurniotu, A. IAPP mimic blocks Aβ cytotoxic self-assembly: cross-suppression of amyloid toxicity of Aβ and IAPP suggests a molecular link between Alzheimer's disease and type II diabetes. Angew. Chem. Int. Ed. 46, 1246–1252 (2007).
Seeliger, J. et al. Cross-amyloid interaction of Aβ and IAPP at lipid membranes. Angew. Chem. Int. Ed. 51, 679–683 (2012).
Ma, B. & Nussinov, R. Selective molecular recognition in amyloid growth and transmission and cross-species barriers. J. Mol. Biol. 421, 172–184 (2012).
Miller, Y., Ma, B. & Nussinov, R. Polymorphism in Alzheimer Aβ amyloid organization reflects conformational selection in a rugged energy landscape. Chem. Rev. 110, 4820–4838 (2010).
Stains, C. I., Mondal, K. & Ghosh, I. Molecules that target beta-amyloid. ChemMedChem 2, 1674–1692 (2007).
Sciarretta, K. L., Gordon, D. J. & Meredith, S. C. Peptide-based inhibitors of amyloid assembly. Methods Enzymol. 413, 273–312 (2006).
Colletier, J-P. et al. Molecular basis for amyloid-β polymorphism. Proc. Natl Acad. Sci. USA 108, 16938–16943 (2011).
Laganowsky, A. et al. Atomic view of a toxic amyloid small oligomer. Science 335, 1228–1231 (2012).
Acknowledgements
The authors acknowledge support from the NIH (5R01 GM049076, 1R01 GM097562 and 1R01 AG029430), the NSF (CHE-1112188, CHE-0750523 and MCB-0445429) and HHMI. The authors also thank A. Berk and D. Gou for help with tissue culture experiments, and S. Blum for suggestions for Fig. 5a.
Author information
Authors and Affiliations
Contributions
P.-N.C., C.L., D.E. and J.S.N. designed the research. P.-N.C., C.L. and M.Z. performed the research. P.-N.C., C.L., M.Z., D.E. and J.S.N. analysed the data. P.-N.C., C.L., M.Z., D.E. and J.S.N. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 15430 kb)
Rights and permissions
About this article
Cite this article
Cheng, PN., Liu, C., Zhao, M. et al. Amyloid β-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity. Nature Chem 4, 927–933 (2012). https://doi.org/10.1038/nchem.1433
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1433
This article is cited by
-
Rationally designed cyclic peptides and nanomaterials as ‘next-generation’ anti-amyloid therapeutics
Journal of Materials Science (2023)
-
Designed peptides as nanomolar cross-amyloid inhibitors acting via supramolecular nanofiber co-assembly
Nature Communications (2022)
-
Mapping the sequence specificity of heterotypic amyloid interactions enables the identification of aggregation modifiers
Nature Communications (2022)
-
Effects of Aβ-derived peptide fragments on fibrillogenesis of Aβ
Scientific Reports (2021)
-
Hyaluronan-carnosine conjugates inhibit Aβ aggregation and toxicity
Scientific Reports (2020)