Abstract
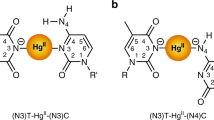
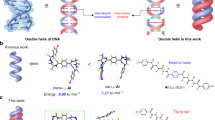
Metal-mediated base pairs represent a powerful tool for the site-specific functionalization of nucleic acids with metal ions. The development of applications of the metal-modified nucleic acids will depend on the availability of structural information on these double helices. We present here the NMR solution structure of a self-complementary DNA oligonucleotide with three consecutive imidazole nucleotides in its centre. In the absence of transition-metal ions, a hairpin structure is adopted with the artificial nucleotides forming the loop. In the presence of Ag(i) ions, a duplex comprising three imidazole–Ag+–imidazole base pairs is formed. Direct proof for the formation of metal-mediated base pairs was obtained from 1J(15N,107/109Ag) couplings upon incorporation of 15N-labelled imidazole. The duplex adopts a B-type conformation with only minor deviations in the region of the artificial bases. This work represents the first structural characterization of a metal-modified nucleic acid with a continuous stretch of metal-mediated base pairs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Niemeyer, C. M. & Mirkin, C. A. Nanobiotechnology (Wiley-VCH, 2004).
Müller, J. Metal-ion-mediated base pairs in nucleic acids. Eur. J. Inorg. Chem. 3749–3763 (2008).
Tanaka, K. & Shionoya, M. Programmable metal assembly on bio-inspired templates. Coord. Chem. Rev. 251, 2732–2742 (2007).
Clever, G. H., Kaul, C. & Carell, T. DNA-metal base pairs. Angew. Chem. Int. Ed. 46, 6226–6236 (2007).
He, W., Franzini, R. M. & Achim, C. Metal-containing nucleic acid structures based on synergetic hydrogen and coordination bonding. Prog. Inorg. Chem. 55, 545–611 (2007).
Weizman, H. & Tor, Y. 2,2'-Bipyridine Ligandoside: A novel building block for modifying DNA with intra-duplex metal complexes. J. Am. Chem. Soc. 123, 3375–3376 (2001).
Meggers, E., Holland, P. L., Tolman, W. B., Romesberg, F. E. & Schultz, P. G. A novel copper-mediated DNA base pair. J. Am. Chem. Soc. 122, 10714–10715 (2000).
Heuberger, B. D., Shin, D. & Switzer, C. Two Watson-Crick-like metallo base-pairs. Org. Lett. 10, 1091–1094 (2008).
Polonius, F.-A. & Müller, J. An artificial base pair, mediated by hydrogen bonding and metal-ion binding. Angew. Chem. Int. Ed. 46, 5602–5604 (2007).
Clever, G. H. & Carell, T. Controlled stacking of 10 transition-metal ions inside a DNA duplex. Angew. Chem. Int. Ed. 46, 250–253 (2007).
Switzer, C., Sinha, S., Kim, P. H. & Heuberger, B. D. A purine-like nickel(ii) base pair for DNA. Angew. Chem. Int. Ed. 44, 1529–1532 (2005).
Tanaka, K., Tengeiji, A., Kato, T., Toyama, N. & Shionoya, M. A discrete self-assembled metal array in artificial DNA. Science 299, 1212–1213 (2003).
Tanaka, K. et al. Programmable self-assembly of metal ions inside artificial DNA duplexes. Nature Nanotech. 1, 190–194 (2006).
Tanaka, Y. et al. 15N–15N J-coupling across HgII: Direct observation of HgII-mediated T–T base pairs in a DNA duplex. J. Am. Chem. Soc. 129, 244–245 (2007).
Ono, A. et al. Specific interactions between Ag(i) ions and cytosine–cytosine pairs in DNA duplexes. Chem. Commun. 4825–4827 (2008).
Wettig, S. D., Wood, D. O., Aich, P. & Lee, J. S. M-DNA: A novel metal ion complex of DNA studied by fluorescence techniques. J. Inorg. Biochem. 99, 2093–2101 (2005).
Johannsen, S., Paulus, S., Düpre, N., Müller, J. & Sigel, R. K. O. Using in vitro transcription to construct scaffolds for one-dimensional arrays of mercuric ions. J. Inorg. Biochem. 102, 1141–1151 (2008).
Schlegel, M. K., Zhang, L., Pagano, N. & Meggers, E. Metal-mediated base pairing within the simplified nucleic acid GNA. Org. Biomol. Chem. 7, 476–482 (2009).
Franzini, R. M. et al. Metal binding to bipyridine-modified PNA. Inorg. Chem. 45, 9798–9811 (2006).
Küsel, A. et al. Metal binding within a peptide-based nucleobase stack with tuneable double-strand topology. Eur. J. Inorg. Chem. 4317–4324 (2005).
Atwell, S., Meggers, E., Spraggon, G. & Schultz, P. G. Structure of a copper-mediated base pair in DNA. J. Am. Chem. Soc. 123, 12364–12367 (2001).
Schlegel, M. K., Essen, L.-O. & Meggers, E. Duplex structure of a minimal nucleic acid. J. Am. Chem. Soc. 130, 8158–8159 (2008).
Böhme, D., Düpre, N., Megger, D. A. & Müller, J. Conformational change induced by metal-ion-binding to DNA containing the artificial 1,2,4-triazole nucleoside. Inorg. Chem. 46, 10114–10119 (2007).
Kuklenyik, Z. & Marzilli, L. G. Mercury(ii) site-selective binding to a DNA hairpin. Relationship of sequence-dependent intra- and interstrand cross-linking to the hairpin–duplex conformational transition. Inorg. Chem. 35, 5654–5662 (1996).
Varani, G., Aboul-ela, F. & Allain, F. H.-T. NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spectrosc. 29, 51–127 (1996).
Tanaka, Y. & Ono, A. Nitrogen-15 NMR spectroscopy of N-metallated nucleic acids: insights into 15N NMR parameters and N–metal bonds. Dalton Trans. 4965–4974 (2008).
Zangger, K. & Armitage, I. M. Silver and gold NMR. Met. Based Drugs 6, 239–245 (1999).
Bowmaker, G. A. et al. Solid-state 109Ag CP/MAS NMR spectroscopy of some diammine silver(i) complexes. Magn. Reson. Chem. 42, 819–826 (2004).
van Stein, G. C., van Koten, G., Vrieze, K., Brevard, C. & Spek, A. L. Structural investigations of silver(i) and copper(i) complexes with neutral N4 donor ligands: X-ray crystal and molecular structure of the dimer [Ag2{μ-(R, S)-1,2-(py-2-CH=N)2Cy}2](O3SCF3)2 and 1H, 13C, and INEPT 109Ag and 15N NMR solution studies. J. Am. Chem. Soc. 106, 4486–4492 (1984).
van Stein, G. C. et al. Group 11 metal ions in poly(donor atom) environments: X-ray crystal and molecular structure of [M((R, S)-1,2-(5-R-thio-2-CH=N)2-c-Hx)2](O3SCF3) (M = Ag(i), R = Me, thio = Thiophene, c-Hx = Cyclohexane) and silver(i) and copper(i) coordination properties in solution (1H, 109Ag, and 15N NMR). Inorg. Chem. 24, 1367–1375 (1985).
Lu, X.-J. & Olson, W. K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 (2003).
Chandrasekaran, R. & Arnott, S. The structure of B-DNA in oriented fibers. J. Biomol. Struct. Dyn. 13, 1015–1027 (1996).
Liu, X. et al. Three novel silver complexes with ligand-unsupported argentophilic interactions and their luminescent properties. Inorg. Chem. 45, 3679–3685 (2006).
Mallajosyula, S. S. & Pati, S. K. Conformational tuning of magnetic interactions in metal–DNA complexes. Angew. Chem. Int. Ed. 48, 4977–4981 (2009).
Gridnev, A. A. & Mihalteva, I. M. Synthesis of 1-alkylimidazoles. Synth. Commun. 24, 1547–1555 (1994).
Rolland, V., Kotera, M. & Lhomme, J. Convenient preparation of 2-deoxy-3,5-di-O-p-toluoyl-α-d-erythro-pentofuranosyl chloride. Synth. Commun. 27, 3505–3511 (1997).
Güntert, P., Mumenthaler, C. & Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program Dyana. J. Mol. Biol. 273, 283–298 (1997).
Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Clore, G. M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003).
Müller, J., Böhme, D., Lax, P., Morell Cerdà, M. & Roitzsch, M. Metal ion coordination to azole nucleosides. Chem. Eur. J. 11, 6246–6253 (2005).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14, 51–55 (1996).
Acknowledgements
Financial support by the European ERAnet-Chemistry, the Swiss National Science Foundation (20EC21-112708 and 200021-124834 to R.K.O.S.), the Swiss State Secretariat for Education and Research (R.K.O.S.), the Deutsche Forschungsgemeinschaft (MU1750/1-3 and MU1750/2-1 to JM), COST D39 and the Fonds der Chemischen Industrie (J.M.) is gratefully acknowledged. We also thank T. van der Wijst for providing us with the partial charges of protonated and unprotonated 1-methylimidazole, as well as R. Micura and K. Breuker, University of Innsbruck, for recording mass spectra.
Author information
Authors and Affiliations
Contributions
J.M. and R.K.O.S. designed research, N.M. and D.B. performed syntheses, S.J. performed the NMR experiments and structure calculations, S.J. and R.K.O.S. analysed data, and J.M., R.K.O.S., S.J. and N.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 627 kb)
Rights and permissions
About this article
Cite this article
Johannsen, S., Megger, N., Böhme, D. et al. Solution structure of a DNA double helix with consecutive metal-mediated base pairs. Nature Chem 2, 229–234 (2010). https://doi.org/10.1038/nchem.512
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.512
This article is cited by
-
Rational design of metal-responsive functional DNA supramolecules
Journal of Inclusion Phenomena and Macrocyclic Chemistry (2024)
-
A paper-based assay for the colorimetric detection of SARS-CoV-2 variants at single-nucleotide resolution
Nature Biomedical Engineering (2022)
-
“Metal-modified base pairs” vs. “metal-mediated pairs of bases”: not just a semantic issue!
JBIC Journal of Biological Inorganic Chemistry (2022)
-
Construction and characterization of metal ion-containing DNA nanowires for synthetic biology and nanotechnology
Scientific Reports (2019)
-
Concerted dynamics of metallo-base pairs in an A/B-form helical transition
Nature Communications (2019)