Abstract
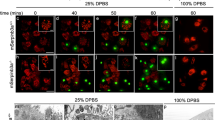
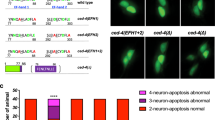
Similar to mammalian excitotoxic cell death, necrotic-like cell death (NCD) in Caenorhabditis elegans can be initiated by hyperactive ion channels. Here we investigate the requirements for genes that execute and regulate programmed cell death (PCD) in necrotic-like neuronal death caused by a toxic MEC-4 channel. Neither the kinetics of necrosis onset nor the total number of necrotic corpses generated is altered by any C. elegans mutation known to block PCD, which provides genetic evidence that the activating mechanisms for NCD and apoptotic cell death are distinct. In contrast, all previously reported ced genes required for phagocytotic removal of apoptotic corpses, as well as ced-12, a new engulfment gene we have identified, are required for efficient elimination of corpses generated by distinct necrosis-inducing stimuli. Our results show that a common set of genes acts to eliminate cell corpses irrespective of the mode of cell death, and provide the first identification of the C. elegans genes that are required for orderly removal of necrotic cells. As phagocytotic mechanisms seem to be conserved from nematodes to humans, our findings indicate that injured necrotic cells in higher organisms might also be eliminated before lysis through a controlled process of corpse removal, a hypothesis that has significant therapeutic implications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hengartner, M. O., Ellis, R. E. & Horvitz, H. R. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356, 494–499 (1992).
Hengartner, M. O. & Horvitz, H. R. C. elegans cell survival gene ced-9 encodes a functional homologue of the mammalian proto-oncogene bcl-2. Cell 76, 665–676 (1994).
Hengartner, M. O. & Horvitz, H. R. Activation of C. elegans cell death protein CED-9 by an amino-acid substitution in a domain conserved in Bcl-2. Nature 369, 318–320 (1994).
Vaux, D. L, Weissman, I. L., & Kim, S. K. Prevention of programmed cell death in Caenorhabditis elegans by human bcl-2. Science 258, 1955–1957 (1992).
Conradt, B. & Horvitz, H. R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93, 519 –529 (1998).
Ellis, H. M. & Horvitz, H. R. Genetic control of programmed cell death in the nematode C. elegans. Cell 44, 817–829 ( 1986).
Yuan, J., Shaham, S., Ledoux, S., Ellis, H. M. & Horvitz, H. R. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1β-converting enzyme. Cell 75, 641–652 (1993).
Xue, D., Shaham S. & Horvitz, H. R. The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 10, 1073–1083 (1996).
Shaham, S. & Horvitz, H. R. An alternatively spliced C. elegans ced-4 RNA encodes a novel cell death inhibitor. Cell 86, 201–208 ( 1996).
Zou, H., Henzel, W. J., Liu, X., Lutschg, A. & Wang, X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405– 413 (1997).
Hedgecock, E. M., Sulston, J. E. & Thomson, J. N. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220, 1277–1279 (1983).
Ellis, R. E., Jacobson, D. M. & Horvitz, H. R. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129, 79–94 ( 1993).
Driscoll, M. & Chalfie, M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature 349, 588–593 (1991).
Hong, K. & Driscoll, M. A transmembrane domain of the putative channel subunit MEC-4 influences mechanotransduction and neurodegeneration in C. elegans. Nature 367, 470– 473 (1994).
Treinin, M. & Chalfie, M. A mutated acetylcholine receptor subunit causes neuronal degeneration in C. elegans. Neuron 14, 871–877 ( 1995).
Treinin, M., Gillo, B. Liebman, L. & Chalfie, M. Two functionally dependent acetylcholine subunits are encoded in a single Caenorhabditis elegans operon. Proc. Natl Acad. Sci. USA 95, 15492–15495 (1998).
Chalfie, M. & Sulston, J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 82, 358–370 ( 1981).
Hall, D. H., et al., Neuropathology of degenerative cell death in Caenorhabditis elegans. J. Neurosci. 17, 1033– 1045 (1992).
Schulze-Osthoff, K. et al. Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. J. Biol. Chem. 267, 5317–5323 (1992).
Dunn, W. A. Autophagy and related mechanisms of lysosome-mediated protein degradation . Trends Cell Biol. 4, 139– 143 (1994).
Kane, D. J., Ord, T., Anton, R. & Bredesen, D. E. Expression of bcl-2 inhibits necrotic neural cell death. J. Neurosci. Res. 40, 269–275 ( 1995).
Vaux, D. L., Whitney, D. & Weismann, I. L. Activation of physiological cell death mechanisms by a necrosis-causing agent. Microsc. Res. Tech. 34 , 259–266 (1996).
Shimizu, S. et al. Retardation of chemical hypoxia-induced necrotic cell death by Bcl-2 and ICE inhibitors: possible involvement of common mediators in apoptotic and necrotic signal transductions. Oncogene 12, 2045–2050 (1996).
Okuno, S. I. et al. Bcl-2 prevents caspase-independent cell death. J. Biol. Chem. 273, 34272–34277 (1998).
Shaham, S. & Horvitz, H. R. Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities . Genes Dev. 10, 578–591 (1996).
Shaham, S., Reddien, P.W., Davies, B., & Horvitz, H.R. Mutational analysis of the C. elegans cell death gene ced-3. Genetics 153, 1655–1671 ( 1999).
Robertson, A. M. G. & Thomson, J. N. Morphology of programmed cell death in the ventral cord of Caenorhabditis elegans . J. Embryol. Exp. Morphol. 67, 89– 100 (1982).
Moynault, A., Luciani, M. F. & Chimini, G. ABC1, the mammalian homologue of the engulfment gene ced-7, is required during phagocytosis of both necrotic and apoptotic cells. Biochem. Soc. Trans. 26, 629– 635 (1998).
Chalfie, M. & Wolinsky, E. The identification and suppression of inherited neurodegeneration in Caenorhabditis elegans . Nature 345, 410–416 (1990).
Korswagen, H. C., Park, J-H., Ohshima, Y., & Plasterk, R. H. A. An activating mutation in a Caenorhabditis elegans Gs protein induces neural degeneration. Genes Dev. 11, 1493 –1503 (1997).
Berger, A. J., Hart, A. C. & Kaplan, J. M. G-α(s)-induced neurodegeneration in Caenorhabditis elegans. J. Neurosci. 18, 2871–2888 (1998).
Wu, Y. C. & Horvitz H. R. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93, 951– 960 (1998).
Reddien, P. W. & Horvitz, H. R. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nature Cell Biol 2, 131– 136 (2000).
Wu, Y. C. & Horvitz, H. R. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180 . Nature 392, 501–504 (1998).
Vercammen, D. et al., Dual signaling of the FAS receptor: initiation of both apoptotic and necrotic cell death pathways. J. Exp. Med. 18, 919–930 (1998).
Kawahara, A., Ohsawa, Y., Matsumura, H., Uchiyama, Y. & Nagata, S. Caspase-independent cell killing by Fas-associated protein with death domain. J. Cell Biol. 143, 1353–1360 (1998).
Chautan, M., Chazal, G., Cecconi, F., Gruss, P. & Goldstein, P. Interdigital cell death can occur through a necrotic and caspase-independent pathway. Curr. Biol. 9, 967–970 (1999).
Brown, S. B., Clarke, M. C. H., Magowan, L., Sanderson, H. & Savill, J. Constitutive death of platelets leading to scavanger-mediated phagocytosis. J. Biol. Chem. 275, 5987–5996 ( 2000).
Savill, J., Fadok, V., Henson, P. & Haslett, C. Phagocyte recognition of cells undergoing apoptosis. Immunol. Today 14, 131–136 (1993).
Hart, S. P., Haslett, C. & Dransfield, I. Recognition of apoptotic cells by phagocytes. Experientia 52, 950–956 (1996).
Fadok, V. A., Bratton, D. Frasch, S. C., Warner, M. L. & Henson, P. H., The role of phosphatidylserine in recognition of apoptotic cells by phagocytes. Cell Death. Diff. 5, 551–562 ( 1998).
Liu, Q. A. & Hengartner, M. O. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans . Cell 93, 961–972 (1998).
Sulston, J. E., Schierenberg, E., White, J. G. & Thomson, J. N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64– 119 (1983).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Sulston, J. E. & Horvitz, H. R. Post embryonic cell lineages of the nematode Caenorhabditis elegans. Dev. Biol. 56, 110–156 ( 1977).
Kimble, J. & Hirsh, D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans . Dev. Biol. 70, 396– 417 (1979).
Driscoll, M. Methods for the study of cell death in the nematode Caenorhabditis elegans . Methods Cell Biol. 46, 323– 353 (1995).
Avery, L. & Horvitz, H. R. A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell 51, 1071– 1078 (1987).
Yuan, J. & Horvitz, H. R. The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development 116, 309–320 ( 1992).
Garcia-Anoveros, J., Ma, C. & Chalfie, M. Regulation of Caenorhabditis elegans degenerin proteins by a putative extracellular domain. Curr. Biol. 5, 441–448 (1995).
Acknowledgements
We thank R. Francis and T. Schedl for providing the oz167 allele. Some strains were provided by The Caenorhabditis Genetics Center, which is supported by the National Institute of Health National Center for Research Resources, M. Chalfie, D. Xue, R. Plasterk and H. R. Horvitz. We thank G. Patterson and members of the Driscoll laboratory for discussion and comments on the manuscript. This work was supported by NIH grants R01NS34435 (to M.D.) and GM-52540 (to M.O.H.); S.C. was supported in part by grant no. 5T32GM08360.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chung, S., Gumienny, T., Hengartner, M. et al. A common set of engulfment genes mediates removal of both apoptotic and necrotic cell corpses in C. elegans. Nat Cell Biol 2, 931–937 (2000). https://doi.org/10.1038/35046585
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/35046585
This article is cited by
-
Engulfing cells promote neuronal regeneration and remove neuronal debris through distinct biochemical functions of CED-1
Nature Communications (2018)
-
ELMOD3, a novel causative gene, associated with human autosomal dominant nonsyndromic and progressive hearing loss
Human Genetics (2018)
-
A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells
Nature Communications (2015)
-
Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells in Caenorhabditis elegans
Nature Cell Biology (2011)
-
Apaf-1-independent programmed cell death in mouse development
Cell Death & Differentiation (2010)