Abstract
The genome is organized via CTCF–cohesin-binding sites, which partition chromosomes into 1–5 megabase (Mb) topologically associated domains (TADs), and further into smaller sub-domains (sub-TADs). Here we examined in vivo an ∼80 kb sub-TAD, containing the mouse α-globin gene cluster, lying within a ∼1 Mb TAD. We find that the sub-TAD is flanked by predominantly convergent CTCF–cohesin sites that are ubiquitously bound by CTCF but only interact during erythropoiesis, defining a self-interacting erythroid compartment. Whereas the α-globin regulatory elements normally act solely on promoters downstream of the enhancers, removal of a conserved upstream CTCF–cohesin boundary extends the sub-TAD to adjacent upstream CTCF–cohesin-binding sites. The α-globin enhancers now interact with the flanking chromatin, upregulating expression of genes within this extended sub-TAD. Rather than acting solely as a barrier to chromatin modification, CTCF–cohesin boundaries in this sub-TAD delimit the region of chromatin to which enhancers have access and within which they interact with receptive promoters.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ghirlando, R. & Felsenfeld, G. CTCF: making the right connections. Genes Dev. 30, 881–891 (2016).
Nichols, M. H. & Corces, V. G. A CTCF code for 3D genome architecture. Cell 162, 703–705 (2015).
Bonev, B. & Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 (2016).
Weth, O. et al. CTCF induces histone variant incorporation, erases the H3K27me3 histone mark and opens chromatin. Nucleic Acids Res. 42, 11941–11951 (2014).
Cuddapah, S. et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19, 24–32 (2008).
Handoko, L. et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 (2011).
Liu, Z., Scannell, D. R., Eisen, M. B. & Tjian, R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146, 720–731 (2011).
Hirayama, T., Tarusawa, E., Yoshimura, Y., Galjart, N. & Yagi, T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2, 345–357 (2012).
Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016).
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016).
Dowen, J. M. et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387 (2014).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Anguita, E., Johnson, C. A., Wood, W. G., Turner, B. M. & Higgs, D. R. Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster. Proc. Natl Acad. Sci. USA 98, 12114–12119 (2001).
Kowalczyk, M. S. et al. Intragenic enhancers act as alternative promoters. Mol. Cell 45, 447–458 (2012).
Hay, D. et al. Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet. 48, 895–903 (2016).
Anguita, E. Deletion of the mouse α-globin regulatory element (HS -26) has an unexpectedly mild phenotype. Blood 100, 3450–3456 (2002).
Davies, J. O. J. et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Meth. 13, 74–80 (2016).
Phillips-Cremins, J. E. et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295 (2013).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Hnisz, D., Day, D. S. & Young, R. A. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 167, 1188–1200 (2016).
Guo, Y. et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162, 900–910 (2015).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA 112, E6456–E6465 (2015).
de Wit, E. et al. CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015).
Rudan, M. V. et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10, 1297–1309 (2015).
Hughes, J. R. et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205–212 (2014).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Narendra, V. et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015).
Yang, R. et al. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 44, 3082–3094 (2016).
Gómez-Marín, C. et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc. Natl Acad. Sci. USA 112, 7542–7547 (2015).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Dekker, J. & Mirny, L. The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 (2016).
Zabidi, M. A. & Stark, A. Regulatory enhancer–core-promoter communication via transcription factors and cofactors. Trends Genet. 32, 801–814 (2016).
Oti, M., Falck, J., Huynen, M. A. & Zhou, H. CTCF-mediated chromatin loops enclose inducible gene regulatory domains. BMC Genom. 17, 252 (2016).
Narendra, V., Bulajić, M., Dekker, J., Mazzoni, E. O. & Reinberg, D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 30, 2657–2662 (2016).
Lower, K. M. et al. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc. Natl Acad. Sci. USA 106, 21771–21776 (2009).
Stadler, M. B. et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495 (2011).
Consortium, T. E. P. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 488, 57–74 (2012).
Simon, C. S. et al. Functional characterisation of cis-regulatory elements governing dynamic Eomes expression in the early mouse embryo. Development 144, 1249–1260 (2017).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Hosseini, M. et al. Causes and consequences of chromatin variation between inbred mice. PLoS Genet. 9, e1003570 (2013).
Leder, A., Daugherty, C., Whitney, B. & Leder, P. Mouse zeta- and alpha-globin genes: embryonic survival, alpha-thalassemia, and genetic background effects. Blood 90, 1275–1282 (1997).
Wallace, H. A. C. et al. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 128, 197–209 (2007).
Spivak, J. L., Toretti, D. & Dickerman, H. W. Effect of phenylhydrazine-induced hemolytic anemia on nuclear RNA polymerase activity of the mouse spleen. Blood 42, 257–266 (1973).
Kina, T. et al. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br. J. Haematol. 109, 280–287 (2000).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Davies, B. et al. Site specific mutation of the Zic2 locus by microinjection of TALEN mRNA in mouse CD1, C3H and C57BL/6J oocytes. PLoS ONE 8, e60216–e60217 (2013).
Cebrian-Serrano, A. et al. Maternal supply of Cas9 to zygotes facilitates the efficient generation of site-specific mutant mouse models. PLoS ONE 12, e0169887–e01698820 (2017).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Nakahashi, H. et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 3, 1678–1689 (2013).
Acknowledgements
The authors would like to thank J. Telenius for her help with bioinformatic analysis of the data and D. Hay, R. Gibbons and A. King for critically reading the manuscript. L.L.P.H. and A.M.O. would like to thank the Wellcome Trust for funding (Chromosome and Developmental Biology PhD Programme, grant code 099684/Z/12/Z; and Wellcome Trust Genomic Medicine and Statistics PhD Programme, grant code 083323/Z/07/Z respectively). The work was further supported by the Wellcome Trust core award 203141/Z/16/Z and the Medical Research Council (MRC Core Funding and Centenary Award reference 4050189188).
Author information
Authors and Affiliations
Contributions
L.L.P.H. planned and carried out experiments, analysed the data, carried out bioinformatic analysis, and wrote the manuscript. M.T.K. coordinated and advised on the project and revised the manuscript. A.M.O. carried out Capture-C experiments and revised the manuscript. M.T.K. and A.M.O. contributed equally to this work. D.B. carried out cell culture and mouse experiments. C.P. carried out mouse microinjection experiments. D.J.D. and M.G. carried out ATAC-seq experiments. J.A.S. and J.A.S.-S. carried out mouse maintenance and haematology experiments. J.R.H. coordinated the project and gave technical and bioinformatics advice throughout the project. B.D. planned and coordinated transgenic mouse experiments and wrote the manuscript. D.R.H. conceived, planned and coordinated the project and wrote the manuscript. B.D. and D.R.H. share senior authorship.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 De novo analysis of the CTCF binding motifs and footprints in mouse erythroid cells.
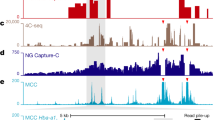
(a) CTCF consensus binding motif in forward and reverse orientation resulting from MEME motif analysis of CTCF ChIP-seq (2 independent experiments in which 2 animals were analysed in total) in erythroid cells. Core (C), upstream (U), and downstream (D) sequence elements are identified with preferential spacing to the core motif (spacer, red bar histograms). (b) Plots of the average DNaseI footprints of C, UC, and UCD motif containing CTCF binding sites in forward orientation (top panel) and reverse orientation (lower panel). Upper (red, +) and lower (blue, -) strand specific DNaseI cleavage signals are shown. Footprints are averaged over the total number of sites in each category (indicated between parentheses). (c) Normalised CTCF ChIP-seq (RPKM, 2 independent experiments in which 2 animals were analysed in total) annotated with the CTCF site names and orientation at the α-globin locus in erythroid cells. Gene annotation is Refseq. DNaseI footprints and top CTCF binding motif hit for each of the CTCF binding sites. Motif P-values are shown (as explained in Fig. 1c). Orientation is indicated by the colour of the arrow over the core motif (C); forward (red) or reverse (blue). Upstream motif (U) is shown in green and downstream motif (D) in yellow.
Supplementary Figure 2 Genome editing strategies for disruption of CTCF binding at the α-globin locus.
Schematic overview of the mutagenesis method (CRISPR-Cas9 and/or TALEN) and the resulting mutations induced at HS-38 (D38), HS-39 (D39), HS-38 and HS-39 (D3839), and HS-29 (D29). DNaseI footprints are annotated with matches to the C, U, and D CTCF motif (coloured bar under C, red = forward, blue = reverse). Where homology directed repair was used to replace part of the CTCF core motif with a restriction site, the single-stranded oligodeoxynucleotides (ssODNs) used as repair templates are shown.
Supplementary Figure 3 Differential interactions of α-globin regulatory regions and flanking genes between wild-type and D29 primary erythroid cells.
Capture-C interaction data for the indicated viewpoints (red vertical bars) in wild-type (WT) and CTCF mutant D29 (deletion of HS-29) primary erythroid cells is shown as described in Fig. 2a. Data represent 3 independent experiments in which 3 animals were analysed in total. Shaded grey bar indicates the mutated CTCF site (HS-29). Also shown are normalised CTCF ChIP-seq (RPKM) for both WT and D29 erythroid cells and ATAC-seq (RPKM) for WT erythroid cells, all merged from 2 independent experiments in which 2 animals were analysed. Gene annotation is Refseq.
Supplementary Figure 4 Effects of the deletion of HS-29 on local gene expression and chromatin state in primary erythroid cells.
(a) MA plot for RNA-seq data derived from WT and D29 erythroid cells. Data represent n = 3 independent experiments in which 3 animals were analysed in total. Mean RNA abundance is plotted on the x-axis and enrichment is plotted on the y-axis. No significant changes in local gene expression were detected as shown on the plot by the genes highlighted in blue: Snrnp25, Rhbdf1, Mpg. Controls are shown in different colours: Mitoferrin-1 (Slc25a37, green), a highly expressed erythroid gene; Sh3pxd2b and Il9r (red), repressed in erythroid; Nprl3 (yellow), a housekeeping gene within the α-globin locus that is unaffected by HS-29 deletion. The five most significant outliers were investigated and only one gene was located in cis to α-globin (Irgm2, >25 Mb removed), making it unlikely that these expression changes represent direct effects from HS-29 deletion. Significance was tested with a Wald test (DEseq2, n = 3 independent experiments in which 3 animals were analysed). (b) Normalised ATAC-Seq and ChIP-seq read-densities (RPKM; 2 independent experiments in which 2 animals were analysed in total) for H3K27me3, H3K4me3, and CTCF at the α-globin locus both in WT and D29 primary erythroid cells. Shaded grey bar indicates HS-29. The dashed box highlights the region over the Rhbdf1 and Mpg genes, magnified in top panels for ease of data visualisation.
Supplementary Figure 5 Effects of the individual deletion of HS-38 or HS-39 on local chromatin state in primary erythroid cells.
Normalised ATAC-seq and ChIP-seq read-densities (RPKM; 2 independent experiments in which 2 animals were analysed in total) for H3K27me3, H3K4me3, and CTCF at the α-globin locus in WT, D38 and D39 primary erythroid cells. Shaded grey bar indicates HS-38 and HS-39 (mutated CTCF sites in D38 and D39). The dashed box highlights the region over the Rhbdf1 and Mpg genes, magnified in top panels for ease of data visualisation.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1721 kb)
Supplementary Information
Supplementary Information (PDF 93 kb)
Supplementary Information
Supplementary Information (XLSX 36 kb)
Rights and permissions
About this article
Cite this article
Hanssen, L., Kassouf, M., Oudelaar, A. et al. Tissue-specific CTCF–cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat Cell Biol 19, 952–961 (2017). https://doi.org/10.1038/ncb3573
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3573
This article is cited by
-
The N-terminal dimerization domains of human and Drosophila CTCF have similar functionality
Epigenetics & Chromatin (2024)
-
Live-cell imaging of chromatin contacts opens a new window into chromatin dynamics
Epigenetics & Chromatin (2023)
-
Enhancer–promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness
Nature Genetics (2023)
-
Transposable elements in mammalian chromatin organization
Nature Reviews Genetics (2023)
-
Capture-C: a modular and flexible approach for high-resolution chromosome conformation capture
Nature Protocols (2022)