Abstract
Autophagy is an adaptation mechanism that is vital for cellular homeostasis in response to various stress conditions. Previous reports indicate that there is a functional interaction between the primary cilium (PC) and autophagy. The PC, a microtubule-based structure present at the surface of numerous cell types, is a mechanical sensor. Here we show that autophagy induced by fluid flow regulates kidney epithelial cell volume in vitro and in vivo. PC ablation blocked autophagy induction and cell-volume regulation. In addition, inhibition of autophagy in ciliated cells impaired the flow-dependent regulation of cell volume. PC-dependent autophagy can be triggered either by mTOR inhibition or a mechanism dependent on the polycystin 2 channel. Only the LKB1–AMPK–mTOR signalling pathway was required for the flow-dependent regulation of cell volume by autophagy. These findings suggest that therapies regulating autophagy should be considered in developing treatments for PC-related diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Satir, P. & Christensen, S. T. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69, 377–400 (2007).
Eggenschwiler, J. T. & Anderson, K. V. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23, 345–373 (2007).
Goetz, S. C. & Anderson, K. V. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11, 331–344 (2010).
Zimmerman, K. & Yoder, B. K. SnapShot: sensing and signaling by cilia. Cell 161, 692 (2015).
Fliegauf, M., Benzing, T. & Omran, H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880–893 (2007).
Hildebrandt, F., Benzing, T. & Katsanis, N. Ciliopathies. N. Engl. J. Med. 364, 1533–1543 (2011).
Abdul-Majeed, S., Moloney, B. C. & Nauli, S. M. Mechanisms regulating cilia growth and cilia function in endothelial cells. Cell Mol. Life Sci. 69, 165–173 (2012).
Kallakuri, S. et al. Endothelial cilia are essential for developmental vascular integrity in zebrafish. J. Am. Soc. Nephrol. 26, 864–875 (2015).
Masyuk, A. I. et al. Cholangiocyte cilia detect changes in luminal fluid flow and transmit them into intracellular Ca2+ and cAMP signaling. Gastroenterology 131, 911–920 (2006).
McGrath, J., Somlo, S., Makova, S., Tian, X. & Brueckner, M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell 114, 61–73 (2003).
Nauli, S. M. et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33, 129–137 (2003).
Weinbaum, S., Duan, Y., Satlin, L. M., Wang, T. & Weinstein, A. M. Mechanotransduction in the renal tubule. Am. J. Physiol. Renal Physiol. 299, F1220–F1236 (2010).
Raghavan, V. & Weisz, O. A. Discerning the role of mechanosensors in regulating proximal tubule function. Am. J. Physiol. Renal Physiol. 310, F1–F5 (2016).
Chapin, H. C. & Caplan, M. J. The cell biology of polycystic kidney disease. J. Cell Biol. 191, 701–710 (2010).
Brown, N. E. & Murcia, N. S. Delayed cystogenesis and increased ciliogenesis associated with the re-expression of polaris in Tg737 mutant mice. Kidney Int. 63, 1220–1229 (2003).
Grantham, J. J., Geiser, J. L. & Evan, A. P. Cyst formation and growth in autosomal dominant polycystic kidney disease. Kidney Int. 31, 1145–1152 (1987).
Boehlke, C. et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat. Cell Biol. 12, 1115–1122 (2010).
Takiar, V. & Caplan, M. J. Polycystic kidney disease: pathogenesis and potential therapies. Biochim. Biophys. Acta 1812, 1337–1343 (2011).
Boya, P., Reggiori, F. & Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 15, 713–720 (2013).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Yang, Z. & Klionsky, D. J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822 (2010).
Kroemer, G., Mariño, G. & Levine, B. Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010).
Meijer, A. J., Lorin, S., Blommaart, E. F. & Codogno, P. Regulation of autophagy by amino acids and MTOR-dependent signal transduction. Amino Acids 47, 2037–2063 (2015).
DuFort, C. C., Paszek, M. J. & Weaver, V. M. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12, 308–319 (2011).
French, A. S. Mechanotransduction. Annu. Rev. Physiol. 54, 135–152 (1992).
He, C. et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481, 511–515 (2012).
Hussain, S. N. et al. Mechanical ventilation-induced diaphragm disuse in humans triggers autophagy. Am. J. Respir. Crit. Care Med. 182, 1377–1386 (2010).
Zaglia, T. et al. Atrogin-1 deficiency promotes cardiomyopathy and premature death via impaired autophagy. J. Clin. Invest. 124, 2410–2424 (2014).
King, J. S. Mechanical stress meets autophagy: potential implications for physiology and pathology. Trends Mol. Med. 18, 583–588 (2012).
Pampliega, O. et al. Functional interaction between autophagy and ciliogenesis. Nature 502, 194–200 (2013).
Tang, Z. et al. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature 502, 254–257 (2013).
Kotsis, F., Boehlke, C. & Kuehn, E. W. The ciliary flow sensor and polycystic kidney disease. Nephrol. Dial. Transplant. 28, 518–526 (2013).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016).
Kimura, S., Noda, T. & Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007).
Heo, J., Sachs, F., Wang, J. & Hua, S. Z. Shear-induced volume decrease in MDCK cells. Cell Physiol. Biochem. 30, 395–406 (2012).
Tanida, I., Minematsu-Ikeguchi, N., Ueno, T. & Kominami, E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1, 84–91 (2005).
Jin, G. et al. Skp2-mediated RagA ubiquitination elicits a negative feedback to prevent amino-acid-dependent mTORC1 hyperactivation by recruiting GATOR1. Mol. Cell 58, 989–1000 (2015).
Orhon, I., Dupont, N., Pampliega, O., Cuervo, A. M. & Codogno, P. Autophagy and regulation of cilia function and assembly. Cell Death Differ. 22, 389–397 (2015).
Wang, S., Livingston, M. J., Su, Y. & Dong, Z. Reciprocal regulation of cilia and autophagy via the MTOR and proteasome pathways. Autophagy 11, 607–616 (2015).
Pazour, G. J. et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151, 709–718 (2000).
Hirokawa, N., Noda, Y., Tanaka, Y. & Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 (2009).
Seglen, P. O. & Gordon, P. B. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl Acad. Sci. USA 79, 1889–1892 (1982).
Briscoe, J. & Therond, P. P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429 (2013).
Hofherr, A. & Kottgen, M. TRPP channels and polycystins. Adv. Exp. Med. Biol. 704, 287–313 (2011).
Jin, X. et al. Cilioplasm is a cellular compartment for calcium signaling in response to mechanical and chemical stimuli. Cell Mol. Life Sci. 71, 2165–2178 (2014).
Cao, M. & Zhong, Q. Cilia in autophagy and cancer. Cilia 5, 4 (2016).
Pampliega, O. & Cuervo, A. M. Autophagy and primary cilia: dual interplay. Curr. Opin. Cell Biol. 39, 1–7 (2016).
Kim, E. S. et al. Inhibition of autophagy suppresses sertraline-mediated primary ciliogenesis in retinal pigment epithelium cells. PLoS ONE 10, e0118190 (2015).
Lam, H. C. et al. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J. Clin. Invest. 123, 5212–5230 (2013).
Meijer, A. J. et al. Cell swelling and the sensitivity of autophagic proteolysis to inhibition by amino acids in isolated rat hepatocytes. Eur. J. Biochem. 215, 449–454 (1993).
vom Dahl, S. et al. Involvement of integrins in osmosensing and signaling toward autophagic proteolysis in rat liver. J. Biol. Chem. 278, 27088–27095 (2003).
Kimura, T. et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J. Am. Soc. Nephrol. 22, 902–913 (2011).
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Chang, T. K. et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat. Cell Biol. 15, 1067–1078 (2013).
Miettinen, T. P. & Bjorklund, M. Mevalonate pathway regulates cell size homeostasis and proteostasis through autophagy. Cell Rep. 13, 2610–2620 (2015).
Fader, C. M., Sanchez, D., Furlan, M. & Colombo, M. I. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9, 230–250 (2008).
Knodler, A. et al. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl Acad. Sci. USA 107, 6346–6351 (2010).
Zeng, H. et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490 (2013).
Huber, T. B. et al. Emerging role of autophagy in kidney function, diseases and aging. Autophagy 8, 1009–1031 (2012).
Ravichandran, K. & Edelstein, C. L. Polycystic kidney disease: a case of suppressed autophagy? Semin. Nephrol. 34, 27–33 (2014).
Wang, Z. & Choi, M. E. Autophagy in kidney health and disease. Antioxid. Redox Signal. 20, 519–537 (2014).
Patel, A. & Honore, E. Polycystins and renovascular mechanosensory transduction. Nat. Rev. Nephrol. 6, 530–538 (2010).
Cebotaru, V. et al. Polycystin-1 negatively regulates Polycystin-2 expression via the aggresome/autophagosome pathway. J. Biol. Chem. 289, 6404–6414 (2014).
Delling, M., DeCaen, P. G., Doerner, J. F., Febvay, S. & Clapham, D. E. Primary cilia are specialized calcium signalling organelles. Nature 504, 311–314 (2013).
Thomas, G. & Hall, M. N. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 9, 782–787 (1997).
Raghavan, V., Rbaibi, Y., Pastor-Soler, N. M., Carattino, M. D. & Weisz, O. A. Shear stress-dependent regulation of apical endocytosis in renal proximal tubule cells mediated by primary cilia. Proc. Natl Acad. Sci. USA 111, 8506–8511 (2014).
Tooze, S. A., Abada, A. & Elazar, Z. Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb. Perspect. Biol. 6, a018358 (2014).
Puri, C., Renna, M., Bento, C. F., Moreau, K. & Rubinsztein, D. C. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154, 1285–1299 (2013).
Dupont, N., Orhon, I., Bauvy, C. & Codogno, P. Autophagy and autophagic flux in tumor cells. Methods Enzymol. 543, 73–88 (2014).
El Boustany, C. et al. Capacitative calcium entry and transient receptor potential canonical 6 expression control human hepatoma cell proliferation. Hepatology 47, 2068–2077 (2008).
Yoo, K. H. et al. Osteopontin regulates renal apoptosis and interstitial fibrosis in neonatal chronic unilateral ureteral obstruction. Kidney Int. 70, 1735–1741 (2006).
Viau, A. et al. Lipocalin 2 is essential for chronic kidney disease progression in mice and humans. J. Clin. Invest. 120, 4065–4076 (2010).
Canaud, G. et al. AKT2 is essential to maintain podocyte viability and function during chronic kidney disease. Nat. Med. 19, 1288–1296 (2013).
Acknowledgements
We thank F. Bienaimé, T. Blanc, C. Nguyen and C. Bauvy for assistance. We also thank T. Yoshimori (Osaka University, Osaksa, Japan) for providing GFP–LC3 and RFP–GFP–LC3 constructs. We are grateful to A. M. Cuervo (Albert Einstein College, Bronx, New York, USA) for sharing KECs and IFT88−/− KECs. We acknowledge the design of the transgenic mouse strains by L. Goldstein, San Diego, California, USA (Kif3atm1Gsn), R. Koesters, Heidelberg, Germany (Tg(Pax8–rtTA2S∗M2)1Koes), and D. Tenen, Boston, USA (Tg(TetOCre)1Dgt). We also acknowledge the Necker Institute Imaging Facility, the Fondation Imagine, the Cochin Imaging Electron Microscopy facility, and the Animal Histology and Morphology Core Facility (SFR Necker INSERM US24, CNRS UMS 3633). The work was supported by institutional funding from INSERM, CNRS, and University Paris Descartes and grants from ANR and INCa to P.C. N.D. is supported by a fellowship from Association pour la Recherche sur le Cancer (ARC), and A.V. was supported by an ERA-EDTA fellowship. E.W.K. is supported by the DFG (KU1504-5/1) and Else–Kröner–Fresenius Stiftung (2011_A87).
Author information
Authors and Affiliations
Contributions
I.O., N.D., M.Z., M.B. and V.B. performed most of the experiments with the exception of electron microscopy experiments, which were performed by A.S. I.B. contributed to the initial part of the project. A.V. and E.W.K. provided the mouse tissues. I.O., N.D., M.Z., T.C., G.F., F.T. and P.C. conceived and planned the experiments and interpreted data. I.O., N.D. and P.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 2 Kinetics of LC3-II and actin expression determined by western blot analyses of WT and IFT88−/− KECs subjected to fluid flow.
(a) WT KEC and (b) IFT88−/− KECs were subjected to flow from 4 h to 4 days (d0, h4, d1, d2, d4) with or without chloroquine (CQ) then LC3-II and actin levels were determined by immunoblotting independently repeated three times. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
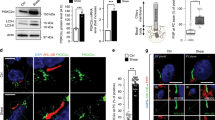
Supplementary Figure 3 Recruitment of ATG16L1 to the basal body on fluid flow induction.
(a–d) WT KEC were subjected to flow or not (d0) for 1 day (d1) then fixed and labelled with γ-tubulin and ATG16L1 antibodies. (a) Cells were analysed by confocal fluorescence microscopy. Arrowheads indicate co-localization of ATG16L1 (green) and γ-tubulin (red). (b) 3D reconstruction of WT KECs subjected to flow or not (dark blue, small cells; white, large cells). (c) Two-fluorescence channel line tracings corresponding to dashed lines in the merge of panel A. (d) Recruitment of ATG16L1 to basal bodies was analysed by quantification of ATG16L1+ basal bodies using ImageJ; n = 125 cells from 5 fields of view per experiment pooled from 3 independent experiments. Data in (d) are means ± s.e.m. Statistical significance was determined using an unpaired Student’s t-test; P values are shown.
Supplementary Figure 4 Ultrastucture analyses of WT and IFT88−/− KECs subjected to fluid flow.
(a–c) WT KEC and IFT88−/− KECs were subjected to flow or not (d0) for 1 day (d1) then fixed and processed for ultrastructure electron microscopy. (a) Representative images. (b) Magnification of double membrane structures (autophagosome) shown in the white inset in panel A. (c) Quantification of autophagic vacuoles (AV) in WT KEC and IFT88−/− KECs with and without (ctrl) flow; n = 15 cells from a single experiment that was independently repeated 3 times. (d) Immunogold electron microscopy for LC3. Data in (c) are means ± s.e.m. Statistical significance was determined using an unpaired Student’s t-test; P values are shown.
Supplementary Figure 5 Effect of fluid flow on ciliogenesis.
(a–d) WT KECs were not subjected to flow (d0) or were subjected to flow for 4 h (h4) or 4 days (d4) then fixed, labelled with acetylated-tubulin (red) and LC3 antibody (green), then analysed by fluorescence microscopy. (a) Representative images of ciliated cells. (b) Number of ciliated cells was quantified using Imaris software. Not significant indicated by ‘ns’. n = 45 cells pooled from 3 independent experiments. (c) Representative images. (d) Cilium length was quantified using Imaris software; n = 45 cells pooled from 3 independent experiments. Data in (b) and (d) are means ± s.e.m. Statistical significance was determined using an unpaired Student’s t-test; P values are shown.
Supplementary Figure 6 Effect of inhibition of ciliogenesis on flow-induced autophagy.
(a) WT KEC and IFT88−/− KECs were subjected to flow or not from 4 h to 4 days (d0, h4, d1, d2, d4). Mean cell volumes were determined by microscopy; ∗, P < 0.05, n = 125 cells from 5 fields of view per experiment pooled from 3 independent experiments. Not significant indicated by ‘ns’. (b–d) KECs engineered to inducibly express an shRNA designed to inhibit expression of kif3a were treated or not with IPTG and subjected to flow for 4 days (d4) or not (d0). (b) Reductions in levels of Kif3a on induction of shRNA expression were confirmed by immunoblotting independently repeated three times. (c) KECs treated or not with IPTG were fixed, labelled with LC3 antibody, and analysed by fluorescence microscopy. Number of endogenous LC3 puncta per cell were quantified using ImageJ software; n = 125 cells from 5 fields of view per experiment pooled from 3 independent experiments. (d,e) KECs engineered to inducibly express shRNA targeting kif3a were treated or not with IPTG and subjected to flow for 4 days (d4) or not (d0). (d) 3D reconstructions of cells (dark blue indicates small cells and white indicates large cells). (e) Mean cell volumes; n = 125 cells from 5 fields of view per experiment pooled from 3 independent experiments. Data in (a), (c), and (e) are means ± s.e.m. Statistical significance was determined using an unpaired Student’s t-test; P values are shown. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
Supplementary Figure 7 Western blot analysis of KECs expressing shRNAs targeting Atg5 or Atg16L1.
Expression of shRNA targeting (a) Atg5 or (b) Atg16L1 in KECs treated or not with IPTG and subjected to flow for 4 days reduced levels of ATG5 and ATG16L1, respectively, as shown by immunoblotting independently repeated three times. (c) KECs engineered to inducibly express shRNA targeting Atg5 were treated or not with IPTG and subjected to flow from 4 h to 4 days (d0, h4, d1, d2, d4), then LC3-II and actin levels were determined by immunoblotting. (d) KECs engineered to inducibly express shRNA targeting Atg16L1 were treated or not with IPTG and subjected to flow from 4 h to 4 days (d0, h4, d1, d2, d4), then LC3-II and actin levels were determined by immunoblotting independently repeated three times. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
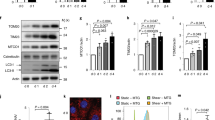
Supplementary Figure 8 Role of Hedgehog signalling and PC2 calcium channel in fluid flow-induced autophagy.
(a) Time course of expression of Gli1 and Ptch1in WT KECs subjected to flow from 4 h to 4 days (4 h, d1, d2, d4) or not (d0). As a positive control, cells were treated with purmophamine for 24 h. Gli1 and Ptch1 mRNA levels were quantified by real-time RT-PCR, normalized with respect to the β-actin gene, and presented as fold increases; from n = 3 independent experiments as shown in i. Not significant indicated by ‘ns’. (b) Cells were subjected to flow for 4 h (h4) or not (d0) or treated with purmophamine for 24 h, then fixed and stained for Smo (green) and acetylated-tubulin (red) to evaluate localization of Smo to the axoneme of cilia. Cells were then analysed by fluorescence microscopy. (c) KECs engineered to inducibly express an shRNA targeting Pkd2 were treated or not with IPTG, and Pkd2 mRNA levels were quantified by real-time RT-PCR, normalized with respect to the β-actin gene, and presented as fold increases; from n = 3 independent experiments as shown in i. (d,e) KECs engineered to inducibly express an shRNA targeting Pkd2 were treated with IPTG (right panel) or not (left panel), labelled with Fura-2, and analysed by fluorescence microscopy. Black arrowheads indicate when cells were subjected to flow or to ATP (to enable evaluation of cell viability). (f,g) KECs engineered to inducibly express an shRNA targeting Pkd2 were treated or not with IPTG and subjected to flow for 4 h or not (d0) with or without CQ. LC3-II/actin ratios visualized by immunoblotting were quantified by densitometry; ∗, P < 0.05, from n = 3 independent experiments as shown in i. (h,i) KECs engineered to inducibly express shRNA targeting Pkd2 were treated or not with IPTG and subjected to flow for 4 days (d4) or not (d0). (h) 3D reconstructions of cells (dark blue indicates small cells and white indicates large cells). (i) Mean cell volumes; n = 125 cells from 5 fields of view per experiment pooled from 3 independent experiments. Data in (a), (c), (g), and (i) are means ± s.e.m. Statistical significance was determined using an unpaired Student’s t-test; P values are shown. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1814 kb)
Rights and permissions
About this article
Cite this article
Orhon, I., Dupont, N., Zaidan, M. et al. Primary-cilium-dependent autophagy controls epithelial cell volume in response to fluid flow. Nat Cell Biol 18, 657–667 (2016). https://doi.org/10.1038/ncb3360
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3360
This article is cited by
-
PKD2/polycystin-2 inhibits LPS-induced acute lung injury in vitro and in vivo by activating autophagy
BMC Pulmonary Medicine (2023)
-
The AMPK-Sirtuin 1-YAP axis is regulated by fluid flow intensity and controls autophagy flux in kidney epithelial cells
Nature Communications (2023)
-
NEK9 regulates primary cilia formation by acting as a selective autophagy adaptor for MYH9/myosin IIA
Nature Communications (2021)
-
Laminar flow inhibits the Hippo/YAP pathway via autophagy and SIRT1-mediated deacetylation against atherosclerosis
Cell Death & Disease (2020)
-
Shear stress turns on the primary cilium and lipophagy
Nature Cell Biology (2020)