Abstract
Age is the primary risk factor for breast cancer in women. Bipotent basal stem cells actively maintain the adult mammary ductal tree, but with age tissues atrophy. We show that cell-extrinsic factors maintain the adult stem cell pool during ageing and dictate tissue stoichiometry. Mammary stem cells spontaneously expand more than 11-fold in virgin adult female mice lacking specific genes for TIMPs, the natural metalloproteinase inhibitors. Compound Timp1/Timp3 null glands exhibit Notch activation and accelerated gestational differentiation. Proteomics of mutant basal cells uncover altered cytoskeletal and extracellular protein repertoires, and we identify aberrant mitotic spindle orientation in these glands, a process that instructs asymmetric cell division and fate. We find that progenitor activity normally declines with age, but enriched stem/progenitor pools prevent tissue regression in Timp mutant mammary glands without affecting carcinogen-induced cancer susceptibility. Thus, improved stem cell content can extend mouse mammary tissue lifespan without altering cancer risk in this mouse model.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Jones, D. L. & Rando, T. A. Emerging models and paradigms for stem cell ageing. Nat. Cell Biol. 13, 506–512 (2011).
Khokha, R. & Werb, Z. Mammary gland reprogramming: metalloproteinases couple form with function. Cold Spring Harb. Perspect. Biol. 3, 1–19 (2011).
Khokha, R., Murthy, A. & Weiss, A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat. Rev. Immunol. 13, 649–665 (2013).
English, J. L. et al. Individual Timp deficiencies differentially impact pro-MMP-2 activation. J. Biol. Chem. 281, 10337–10346 (2006).
Fata, J. E., Chaudhary, V. & Khokha, R. Cellular turnover in the mammary gland is correlated with systemic levels of progesterone and not 17β-estradiol during the estrous cycle. Biol. Reprod. 65, 680–688 (2001).
Shimoda, M. et al. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat. Cell Biol. 16, 889–901 (2014).
Pike, M. C., Krailo, M. D., Henderson, B. E., Casagrande, J. T. & Hoel, D. G. “Hormonal” risk factors, “breast tissue age” and the age-incidence of breast cancer. Nature 303, 767–770 (1983).
Asselin-Labat, M-L. et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201–209 (2007).
Shehata, M. et al. Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 14, R134 (2012).
Santagata, S. et al. Taxonomy of breast cancer based on normal cell phenotype predicts outcome. J. Clin. Invest. 124, 859–870 (2014).
Badders, N. M. et al. The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS ONE 4, e6594 (2009).
Teulière, J. et al. Targeted activation of β-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development 132, 267–277 (2005).
Joseph, H., Gorska, A. E., Sohn, P., Moses, H. L. & Serra, R. Overexpression of a kinase-deficient transforming growth factor-β type II receptor in mouse mammary stroma results in increased epithelial branching. Mol. Biol. Cell 10, 1221–1234 (1999).
Amour, A. et al. The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 473, 275–279 (2000).
Amour, A. et al. TNF-α converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 435, 39–44 (1998).
Gibb, D. R. et al. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J. Exp. Med. 207, 623–635 (2010).
Murthy, A. et al. Notch activation by the metalloproteinase ADAM17 regulates myeloproliferation and atopic barrier immunity by suppressing epithelial cytokine synthesis. Immunity 36, 105–119 (2012).
Raafat, A. et al. Expression of Notch receptors, ligands, and target genes during development of the mouse mammary gland. J. Cell. Physiol. 226, 1940–1952 (2011).
Andersson, E. R., Sandberg, R. & Lendahl, U. Notch signaling: simplicity in design, versatility in function. Development 138, 3593–3612 (2011).
Lee, C. W. et al. A functional Notch-survivin gene signature in basal breast cancer. Breast Cancer Res. 10, R97 (2008).
Bouras, T. et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 3, 429–441 (2008).
Raouf, A. et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell 3, 109–118 (2008).
Rios, A. C., Fu, N. Y., Lindeman, G. J. & Visvader, J. E. In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322–327 (2014).
Oakes, S. R. et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 22, 581–586 (2008).
Meier-Abt, F. et al. Parity induces differentiation and reduces Wnt/Notch signaling ratio and proliferation potential of basal stem/progenitor cells isolated from mouse mammary epithelium. Breast Cancer Res. 15, R36 (2013).
Linder, S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 17, 107–117 (2007).
Taddei, I. et al. β1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat. Cell Biol. 10, 716–722 (2008).
Regan, J. L. et al. Aurora A kinase regulates mammary epithelial cell fate by determining mitotic spindle orientation in a Notch-dependent manner. Cell Rep. 4, 110–123 (2013).
Neumüller, R. A & Knoblich, J. A. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 23, 2675–2699 (2009).
Klein, A. M. & Simons, B. D. Universal patterns of stem cell fate in cycling adult tissues. Development 138, 3103–3111 (2011).
Garbe, J. C. et al. Accumulation of multipotent progenitors with a basal differentiation bias during aging of human mammary epithelia. Cancer Res. 72, 3687–3701 (2012).
Flurkey, K., Currer, J. M. & Harrison, D. E. in The Mouse in Biomedical Research Vol. 3, 637–672 (Elsevier, 2007).
Ghosh, K. et al. Association between mammographic density and age-related lobular involution of the breast. J. Clin. Oncol. 28, 2207–2212 (2010).
Shoker, B. S., Jarvis, C., Sibson, D. R., Walker, C. & Sloane, J. P. Oestrogen receptor expression in the normal and pre-cancerous breast. J. Pathol. 188, 237–244 (1999).
Sprenger, C. C., Plymate, S. R. & Reed, M. J. Aging-related alterations in the extracellular matrix modulate the microenvironment and influence tumor progression. Int. J. Cancer 127, 2739–2748 (2010).
Rosen, P. P. Rosen’s Breast Pathology 29–32 (Lippincott-Raven, 1997).
Kessenbrock, K., Plaks, V. & Werb, Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141, 52–67 (2010).
Geigl, J. B. et al. Analysis of gene expression patterns and chromosomal changes associated with aging. Cancer Res. 64, 8550–8557 (2004).
Ly, D. H., Lockhart, D. J., Lerner, R. A. & Schultz, P. G. Mitotic misregulation and human aging. Science 287, 2486–2492 (2000).
Glukhova, M. A & Streuli, C. H. How integrins control breast biology. Curr. Opin. Cell Biol. 25, 633–641 (2013).
Streuli, C. H. Integrins and cell-fate determination. J. Cell Sci. 122, 171–177 (2009).
LaBarge, M. A. et al. Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments. Integr. Biol. (Camb.) 1, 70–79 (2009).
El-Hashash, A. H. K. et al. Eya1 controls cell polarity, spindle orientation, cell fate and Notch signaling in distal embryonic lung epithelium. Development 138, 1395–1407 (2011).
Das, R. M. & Storey, K. G. Mitotic spindle orientation can direct cell fate and bias Notch activity in chick neural tube. EMBO Rep. 13, 448–454 (2012).
Williams, S. E., Beronja, S., Pasolli, H. A. & Fuchs, E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 470, 353–358 (2011).
Conboy, I. M., Conboy, M. J., Smythe, G. M. & Rando, T. A. Notch-mediated restoration of regenerative potential to aged muscle. Science 302, 1575–1577 (2003).
Tomasetti, C. & Vogelstein, B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Nature 347, 78–81 (2015).
Simons, B. D. & Clevers, H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell 145, 851–862 (2011).
Martins, V. C. et al. Cell competition is a tumour suppressor mechanism in the thymus. Nature 509, 465–470 (2014).
Signer, R. A. J. & Morrison, S. J. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell 12, 152–165 (2013).
Lee, J. A., Chin, P. G., Kukull, W. A., Tompkins, R. S. & Weatherall, A. F. Relationship of age to incidence of breast cancer in young women. J. Natl Cancer Inst. 57, 753–756 (1976).
Joshi, P. A. et al. Progesterone induces adult mammary stem cell expansion. Nature 465, 1–5 (2010).
Schramek, D. et al. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature 468, 98–102 (2010).
Aldaz, C. M., Liao, Q. Y., LaBate, M. & Johnston, D. A. Medroxyprogesterone acetate accelerates the development and increases the incidence of mouse mammary tumors induced by dimethylbenzanthracene. Carcinogenesis 17, 2069–2072 (1996).
Cardiff, R. D. et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene 19, 968–988 (2000).
Sinha, A., Ignatchenko, V., Ignatchenko, A., Mejia-Guerrero, S. & Kislinger, T. In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochem. Biophys. Res. Commun. 445, 694–701 (2014).
Acknowledgements
The authors thank Marco A. Di Grappa, S. R. Narala and H. Fang for technical assistance, Cedric Blanpain (Brussels, Belgium) for providing the wholemount immunofluorescence protocol and P. Joshi for critical review of the manuscript. H.W.J. was awarded a Canadian Breast Cancer Foundation doctoral fellowship, and this research was supported by funding from the Canadian Breast Cancer Foundation and the Canadian Cancer Society Research Institute to R.K.
Author information
Authors and Affiliations
Contributions
H.W.J. and R.K. designed the study and wrote the manuscript; H.W.J. and P.W. carried out limiting dilution assay cell transplants; A.S. and T.K. carried out mass spectrometry proteomics; H.K.B. directed mitosis scoring, advised on mammary tissue ageing and classified mammary tumours; H.W.J. carried out and analysed all other experiments; R.K. directed the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 4 FACS analysis of luminal and basal cell populations across the family of Timp deficient mammary glands and with age in Timp1−/−/Timp3−/− glands.
a, Representative FACS plots of luminal (CD24+CD49flo) and basal (CD24+CD49fhi) populations generated from single and compound Timp mutant glands as indicated. b, Total lin−(ter119−CD45−CD31−) mammary cell number after collagenase digestion from individual and compound 6-month-old Timp deficient mammary glands (mean ± s.e.m., one-way ANOVA; n = 3 WT, 4 T1, 4 T3, 5 T1T3 ∗∗∗P < 0.0001, 4 T1T2T4 mice). c, Quantification of total luminal (red), basal (blue) and stromal (grey) (CD24−CD49f−) populations in 6-month-old adult mammary glands (mean ± s.e.m., one-way ANOVA; n = 3 WT, 4 T1, 4 T3, 5 T1T3, 4 T1T2T4 mice; Basal T1T3 p = 0.0002, Stromal T1T3 p = 0.0378). Solid bars represent Timp1−/−/Timp3−/− glands. d, Quantification of total luminal (CD24+CD49flo), basal (CD24+CD49fhi) and stromal (CD24−CD49f−) in 9-week or 6-month old WT or Timp1−/−/Timp3−/− mice, as well as their further segregated luminal differentiated (CD49floCD61+), luminal progenitor (CD49floCD61+) and basal (CD49fhiCD61+) (e) cell populations (mean ± s.e.m., two-way ANOVA with Tukey’s multiple comparisons test; 9 weeks n = 3 WT, 7 T1T3 mice and 6 months n = 6 WT, 5 T1T3 mice ∗p = 0.0400,∗∗∗P < 0.0001). f, Representative FACS plots of WT and Timp1−/−/Timp3−/− Sca1 and CD49b stained luminal cells. g, Enumeration of total Sca1 and CD49b segregated luminal cells as well as basal (CD24+ CD49fhi, blue) cells in 6-month-old mammary glands (mean ± s.e.m., Student’s T-test; n = 3, Luminal Progenitor p = 0.0080, Basal p = 0.0504). h, Measurement of serum hormones in 6-month-old WT and Timp1−/−/Timp3−/− mice (mean ± s.e.m., Student’s T-test; n = 4 WT, 3 T1T3 mice).
Supplementary Figure 5 Comparison of cell type specific marker gene expression from total gland and FACS-purified cell populations.
a, qRT-PCR expression in total mammary homogenates (black, mean ± s.e.m., n = 3 mice) and flow cytometry sorted luminal differentiated (CD49floCD61−, yellow), luminal progenitor (CD49floCD61+, red), basal (CD24+CD49fhiCD61+, blue) and stromal (CD24−CD49f−, grey) cell populations (coloured, mean ± s.e.m., n = 4 WT, 6 T1T3 mice). Cell markers used include ER & PR for luminal differentiated hormone receptor positive cells, Keratin 8, Keratin 18 for luminal cells, Keratin 5, Keratin 14 & p63 for basal cells and Fibroblast activated protein (FAP) for stromal fibroblasts. b, Representative immunofluorescence image of PR (red), Keratin 5 (green) and DAPI (blue) completed in n = 3 mice (scale bar 10 μm).
Supplementary Figure 6 Wnt and TGFβ signalling in WT and Timp1−/−/Timp3−/− mammary glands.
a, Quantification of Wnt target genes cMyc and TCF7 by qRT-PCR in FACS-purified cell populations (luminal differentiated, CD49floCD61+, yellow; luminal progenitor, CD49floCD61+, red; basal, CD24+CD49fhiCD61+, red; stromal, CD24−CD49f−, grey; mean ± s.e.m., two-way ANOVA with Šidák’s multiple comparisons test; n = 3 mice). b, Representative immunofluorescent images of β-catenin (red), Keratin 5 (green), and DAPI nuclear stain (blue) in WT and Timp1−/−/Timp3−/− mammary glands (n = 3 mice, scale bar 20 μm (top) and 5 μm (bottom). c, Immunoblotting of total mammary gland lysates for unphosphorylated (active) β-catenin, total β-catenin and β-actin as loading control; d, qRT-PCR expression of TGFβ target gene Wnt5a and TGFβ1 ligand in FACS-purified cell populations (mean ± s.e.m., two-way ANOVA with Šidák’s multiple comparisons test; n = 3 mice). e, Immunoblots of phosphorylated SMAD2/3, total SMAD and loading control β-actin. f, ELISA for activated TGFβ ligand per 150 μg of total mammary gland lysate (mean ± s.e.m., Student’s T-test; n = 3 WT, 4 T1T3 mice).
Supplementary Figure 7 Mammary luminal cell populations during gestation and after involution or ovariectomy.
Representative FACS-plots and quantification (mean ± s.e.m., Student’s T-test) of luminal differentiated (CD49floCD61−, yellow), and luminal progenitor (CD49fhiCD61+, red) (a,c,e), or hormone receptor positive (CD49b−Sca1+, yellow) and luminal progenitor (CD49b+Sca1−, red) populations (b,d,f) in WT and Timp1−/−/Timp3−/− adult mammary glands at 16.5 dpc (ab; n = 3 WT, 4 T1T3 mice), post-involution (c,d; n = 3 mice), or after ovariectomy (e,f; n = 5 WT, 4 T1T3 mice).
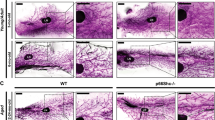
Supplementary Figure 8 Unaltered integrin levels, increased gelatinase activity and divergent mitotic orientation in Timp1−/−/Timp3−/− mammary glands.
a, Protein intensity comparison of integrin family proteins detected by mass spectrometry (mean ± s.e.m., Student’s T-test; n = 3 mice). b, Gelatin zymography of mammary tissue lysates indicates increased MMP9 as well as increased mature/active MMP2 in Timp1−/−/Timp3−/−glands. c, H&E images of representative mitotic orientations, scale bar 20 μm. d, Radial histograms of mitotic spindle angles relative to the adjacent tissue boundary (lumen or matrix). e, Histograms of the average absolute number of mitosis per 1 mm2 grouped by angle of division per genotype (mean ± s.e.m., two-way ANOVA with Šidák’s multiple comparisons test; n = 5 mice). f, Quantification of luminal (red) or basal (blue) mitosis as a percentage of total mammary ductal cells in mice >6 months of age (mean ± s.e.m., Student’s T-test; n = 5 mice, ∗p = 0.0140) g, Representative immunofluorescence of phosho-histone H3 positive mitotic DNA (green) and the SMA positive basal compartment (red) completed in n = 10 mice; scale bar 20 μm, inset scale bar 5 μm.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1748 kb)
Supplementary Video 1
Z stack image of basal Timp1−/−/Timp3−/− mitotic cell. Movie of rotating confocal microscopy Z stack of immunofluorescence stained Timp1−/−/Timp3−/− mammary gland. DAPI (blue), SMA (red), and phospho-Histone H3 (green). (AVI 73 kb)
Rights and permissions
About this article
Cite this article
Jackson, H., Waterhouse, P., Sinha, A. et al. Expansion of stem cells counteracts age-related mammary regression in compound Timp1/Timp3 null mice. Nat Cell Biol 17, 217–227 (2015). https://doi.org/10.1038/ncb3118
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3118
This article is cited by
-
mTOR inhibition abrogates human mammary stem cells and early breast cancer progression markers
Breast Cancer Research (2023)
-
Identifying the murine mammary cell target of metformin exposure
Communications Biology (2019)
-
TIMPs: versatile extracellular regulators in cancer
Nature Reviews Cancer (2017)
-
Cellular mechano-environment regulates the mammary circadian clock
Nature Communications (2017)