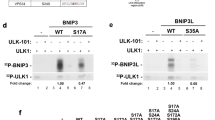
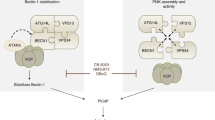
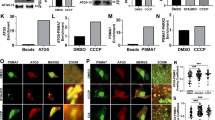
Abstract
Autophagy is important in the basal or stress-induced clearance of bulk cytosol, damaged organelles, pathogens and selected proteins by specific vesicles, the autophagosomes. Following mTOR (mammalian target of rapamycin) inhibition, autophagosome formation is primed by the ULK1 and the beclin-1–Vps34–AMBRA1 complexes, which are linked together by a scaffold platform, the exocyst. Although several regulative steps have been described along this pathway, few targets of mTOR are known, and the cross-talk between ULK1 and beclin 1 complexes is still not fully understood. We show that under non-autophagic conditions, mTOR inhibits AMBRA1 by phosphorylation, whereas on autophagy induction, AMBRA1 is dephosphorylated. In this condition, AMBRA1, interacting with the E3-ligase TRAF6, supports ULK1 ubiquitylation by LYS-63-linked chains, and its subsequent stabilization, self-association and function. As ULK1 has been shown to activate AMBRA1 by phosphorylation, the proposed pathway may act as a positive regulation loop, which may be targeted in human disorders linked to impaired autophagy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2012).
Hosokawa, N. et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 20, 1981–1991 (2009).
Chan, E. Y., Longatti, A., McKnight, N. C. & Tooze, S. A. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Biol. Cell. 29, 157–171 (2009).
Jung, C. H. et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009).
Hara, T. et al. FIP200, an ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 (2008).
Hokosawa, N. et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5, 973–979 (2009).
Ganley, I. G. et al. ULK1–ATG13-FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 (2009).
Mercer, C. A., Kaliappan, A. & Dennis, P. B. A novel, human Atg13 binding protein, Atg101, interacts with Ulk1 and is essential for macroautophagy. Autophagy 5, 649–662 (2009).
Hayashi-Nishino, M. et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 (2009).
Fimia, G. M. et al. Ambra1 regulates autophagy and development of the nervous system. Nature 447, 1121–1125 (2007).
Di Bartolomeo, S. et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 191, 155–168 (2010).
Chan, E. Y., Kir, S. & Tooze, S. A. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282, 25464–25474 (2007).
Chen, Z. J. & Sun, L. J. Non proteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 (2009).
Hoeller, D. & Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 (2009).
Raiborg, C. & Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–445 (2009).
Zhou, X. et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc. Natl Acad. Sci. USA 104, 5842–5847 (2007).
Shi, C. S. & Kehrl, J. H. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Sci. Signal 3, 123 (2010).
Ye, H. et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418, 443–447 (2002).
Liu, Z., Zhang, W. P., Xing, Q., Ren, X., Liu, M. & Tang, C. Noncovalent dimerization of ubiquitin. Angew. Chem. Int. Ed. Engl. 51, 469–472 (2012).
Humphrey, R. K., Yu, S. M., Bellary, A., Gonuguntla, S., Yebra, M. & Jhala, U. S. Lysine 63-linked ubiquitination modulates mixed lineage kinase-3 interaction with JIP1 scaffold protein in cytokine-induced pancreatic β cell death. J. Biol. Chem. 288, 2428–2440 (2012).
Yeh, Y. Y., Shah, K. H. & Herman, P. K. An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J. Biol. Chem. 286, 28931–28939 (2011).
Hsu, P. P. et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 (2011).
Feldman, M. E. et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38 (2009).
Thoreen, C. C. et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. Biol. Chem. 284, 8023–8032 (2009).
Itakura, E. & Mizushima, N. Characterization of autophagosome formation siteby a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776 (2010).
Bodemann, B. O. et al. RalB and the exocyst mediate the cellular starvationresponse by direct activation of autophagosome assembly. Cell 144, 253–267 (2011).
Hosking, R. mTOR: the master regulator. Cell 149, 955–957 (2012).
Dyson, H. J. & Wright, P. E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 253–267 (2005).
Sorrentino, A. et al. The type I TGF- β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 10, 1199–1207 (2008).
Jin, J., Arias, E. E., Chen, J., Harper, J. W. & Walter, J. C. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 (2006).
Behrends, C., Sowa, M. E., Gygi, S. P. & Harper, J. W. Network organization of the human autophagy system. Nature 466, 68–76 (2010).
Takaesu, G. et al. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell 5, 649–658 (2000).
Wertz, I. E. et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 (2004).
Tomoda, T., Bhatt, R. S., Kuroyanagi, H., Shirasawa, T. & Hatten, M. E. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24, 833–846 (1999).
Acknowledgements
We thank the Animal Facility (STA) of the University of Rome ‘Tor Vergata’ for the mouse work, M. Acuña Villa and M. W. Bennett for editorial and secretarial work, and G. Basile and M. Corrado for research assistance. We are indebted to S. A. Tooze (Cancer Research Institute London, UK), D. H. Kim (University of Minnesota Cancer Center, USA), and D. M. Sabatini and R. Zoncu (Whitehead Institute for Biomedical Research, USA) for kindly providing us with ULK1 and ATG13 constructs and HEK293 cells stably expressing RAPTOR–FLAG, respectively. This work was supported in part by grants from the Telethon Foundation (GGP10225), AIRC (IG2010 and IG2012 to FC and MP), FISM (2009), the Italian Ministry of University and Research (PRIN 2009 and FIRB Accordi di Programma 2011) and the Italian Ministry of Health (Ricerca Finalizzata and Ricerca Corrente to F.C., M.P. and G.M.F.).
Author information
Authors and Affiliations
Contributions
F.N. performed most experiments with crucial help from F.S. (immunofluorescence and confocal analyses), M.A. (mutagenesis and cloning), V.C. (immunoprecipitation analysis), M.B. (real-time PCR) and P.B. (kinase assay in vitro). C.G. and J.D. performed the mass spectrometry analysis; G.M.F. provided critical reagents. F.N. and F.C. wrote the manuscript with the help and suggestions of G.M.F. and M.P.; F.C. conceived and designed the research. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2635 kb)
Supplementary Table 1
Supplementary Information (XLS 59 kb)
Rights and permissions
About this article
Cite this article
Nazio, F., Strappazzon, F., Antonioli, M. et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 15, 406–416 (2013). https://doi.org/10.1038/ncb2708
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2708
This article is cited by
-
Gut microbial metabolites SCFAs and chronic kidney disease
Journal of Translational Medicine (2024)
-
Zebrafish ambra1b knockout reveals a novel role for Ambra1 in primordial germ cells survival, sex differentiation and reproduction
Biological Research (2023)
-
The role of NEDD4 related HECT-type E3 ubiquitin ligases in defective autophagy in cancer cells: molecular mechanisms and therapeutic perspectives
Molecular Medicine (2023)
-
Cezanne promoted autophagy through PIK3C3 stabilization and PIK3C2A transcription in lung adenocarcinoma
Cell Death Discovery (2023)
-
The deubiquitinase Leon/USP5 interacts with Atg1/ULK1 and antagonizes autophagy
Cell Death & Disease (2023)